3QYA
 
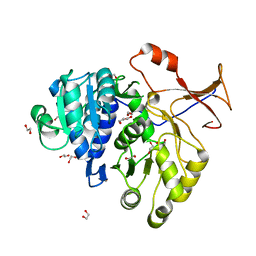 | | Crystal structure of a red-emitter mutant of Lampyris turkestanicus luciferase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kheirabadi, M, Gohlke, U, Hossein Khani, S, Heinemann, U, Naderi-Manesh, H. | | Deposit date: | 2011-03-03 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of native and a mutant of Lampyris turkestanicus luciferase implicate in bioluminescence color shift.
Biochim.Biophys.Acta, 1834, 2013
|
|
4TPJ
 
 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | ALA-ALA-ALA, DODECYL-BETA-D-MALTOSIDE, Proton:oligopeptide symporter POT family, ... | | Authors: | Guettou, F, Quistgaard, E, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4TPH
 
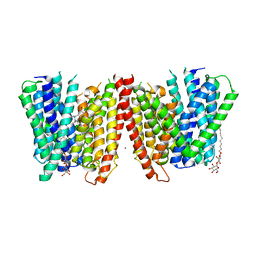 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | 3,5 DIBROMOTYROSINE, ALANINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Guettou, F, Quistgaard, E, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4TPG
 
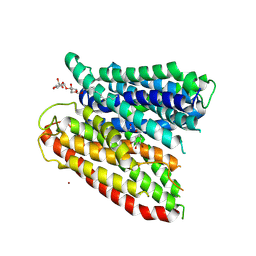 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | Ala-L-3-Br-Tyr-Ala, DODECYL-BETA-D-MALTOSIDE, Proton:oligopeptide symporter POT family, ... | | Authors: | Guettou, F, Quistgaard, E.M, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6C8H
 
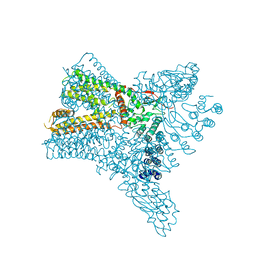 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of gadolinium | | Descriptor: | GADOLINIUM ATOM, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C8G
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of barium | | Descriptor: | BARIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C8F
 
 | | Crystal structure of Transient Receptor Potential (TRP) channel TRPV4 in the presence of cesium | | Descriptor: | CESIUM ION, Transient receptor potential cation channel, subfamily V, ... | | Authors: | Deng, Z, Paknejad, N, Maksaev, G, Sala-Rabanal, M, Nichols, C.G, Hite, R.K, Yuan, P. | | Deposit date: | 2018-01-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2YR6
 
 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | Descriptor: | 2-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR5
 
 | | Crystal structure of L-phenylalanine oxidase from Psuedomonas sp.P501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Pro-enzyme of L-phenylalanine oxidase, ... | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Hikima, T, Yamamoto, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
2YR4
 
 | | Crystal structure of L-phenylalanine oxiase from Psuedomonas sp. P-501 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pro-enzyme of L-phenylalanine oxidase, SULFATE ION | | Authors: | Ida, K, Kurabayashi, M, Suguro, M, Suzuki, H. | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501.
J.Biol.Chem., 283, 2008
|
|
4M46
 
 | |
6YLC
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | 5-fluoranyl-2-[5-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl]benzenecarbonitrile, Mitogen-activated protein kinase 6 | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6YKY
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | 3-(4-methoxyphenyl)-~{N}-[(3~{R})-1-pyridin-4-ylpyrrolidin-3-yl]-[1,2,3]triazolo[4,5-d]pyrimidin-5-amine, Mitogen-activated protein kinase 6 | | Authors: | Graedler, U. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6YLL
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | Mitogen-activated protein kinase 6, ~{N}4-[3-(4-methoxyphenyl)-[1,2,3]triazolo[4,5-d]pyrimidin-5-yl]cyclohexane-1,4-diamine | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7KNW
 
 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
8FYG
 
 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8FNX
 
 | | Crystal structure of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | GLYCEROL, Hyaluronate lyase, PHOSPHATE ION | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2022-12-28 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
8G0O
 
 | | Crystal structure of Y281F mutant of Hyaluronate lyase B from Cutibacterium acnes | | Descriptor: | Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
5Y31
 
 | | Crystal structure of human LGI1-ADAM22 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 22, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (7.125 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
5Y30
 
 | | Crystal structure of LGI1 LRR domain | | Descriptor: | Leucine-rich glioma-inactivated protein 1 | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
5Y2Z
 
 | | Crystal structure of human LGI1 EPTP-ADAM22 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of epilepsy-related ligand-receptor complex LGI1-ADAM22.
Nat Commun, 9, 2018
|
|
7STF
 
 | | Structure of KRAS G12V/HLA-A*03:01 in complex with antibody fragment V2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Wright, K.M, Gabelli, S.B, Miller, M. | | Deposit date: | 2021-11-12 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Hydrophobic interactions dominate the recognition of a KRAS G12V neoantigen.
Nat Commun, 14, 2023
|
|
5ME8
 
 | | N-terminal domain of the human tumor suppressor ING5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Inhibitor of growth protein 5 | | Authors: | Roversi, P, Blanco, F.J, Rojas, A.L, Buitrago, J.A.R. | | Deposit date: | 2016-11-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
5MTO
 
 | | N-terminal domain of the human tumor suppressor ING5 C19S mutant | | Descriptor: | Inhibitor of growth protein 5, SODIUM ION, SULFATE ION | | Authors: | Ormaza, G, Buitrago, J.A.R, Roversi, P, Rojas, A.L, Blanco, F.J. | | Deposit date: | 2017-01-10 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Tumor Suppressor ING5 Is a Dimeric, Bivalent Recognition Molecule of the Histone H3K4me3 Mark.
J.Mol.Biol., 431, 2019
|
|
