6OVE
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-propylphenyl-ACEPC | | Descriptor: | (3R,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(4-propylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
6OVD
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC | | Descriptor: | (3S,5S)-5-[(2R)-2-amino-2-carboxyethyl]-1-(3-ethylphenyl)pyrazolidine-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Syrenne, J.T, Mou, T.C, Tamborini, L, Pinto, A, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 3-ethylphenyl-ACEPC
To Be Published
|
|
3DTP
 
 | | Tarantula heavy meromyosin obtained by flexible docking to Tarantula muscle thick filament Cryo-EM 3D-MAP | | Descriptor: | Myosin II regulatory light chain, Myosin light polypeptide 6, Myosin-11,Myosin-7 | | Authors: | Alamo, L, Wriggers, W, Pinto, A, Bartoli, F, Salazar, L, Zhao, F.Q, Craig, R, Padron, R. | | Deposit date: | 2008-07-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Three-Dimensional Reconstruction of Tarantula Myosin Filaments Suggests How Phosphorylation May Regulate Myosin Activity
J.Mol.Biol., 384, 2008
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
3JAX
 
 | | Heavy meromyosin from Schistosoma mansoni muscle thick filament by negative stain EM | | Descriptor: | myosin 2 heavy chain, myosin regulatory light chain, smooth muscle myosin essential light chain | | Authors: | Sulbaran, G, Alamo, L, Pinto, A, Marquez, G, Mendez, F, Padron, R, Craig, R. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | An invertebrate smooth muscle with striated muscle myosin filaments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VIH
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-fluorophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-fluorophenyl)-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCINE, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VIJ
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-bromophenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-(4-bromophenyl)-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VII
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-(3-fluoropropyl)phenyl-ACEPC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DEX
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, phenyl-ACEPC | | Descriptor: | 5-[(2R)-2-amino-2-carboxyethyl]-1-phenyl-1H-pyrazole-3-carboxylic acid, GLYCINE, Glutamate receptor ionotropic, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 2017
|
|
6FFM
 
 | |
8Q7K
 
 | | IRGQ LIR2 peptide in complex with LC3B | | Descriptor: | 1,2-ETHANEDIOL, Immunity-related GTPase family Q protein, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Misra, M, Bailey, H.J, Pomirska, J, Dikic, I. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tumor immune evasion through IRGQ-directed autophagy
To Be Published
|
|
6YC8
 
 | | Crystal structure of KRED1-Pglu enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Di Pisa, F. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into the desymmetrization of bulky 1,2-dicarbonyls through enzymatic monoreduction.
Bioorg.Chem., 108, 2021
|
|
4CJX
 
 | | The crystal structure of Trypanosoma brucei N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, C-1-TETRAHYDROFOLATE SYNTHASE, CYTOPLASMIC, ... | | Authors: | Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-12-23 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of 2,4-Diamino-6-Oxo-1,6-Dihydropyrimidin-5-Yl Ureido Based Inhibitors of Trypanosoma Brucei Fold and Testing for Antiparasitic Activity.
J.Med.Chem., 58, 2015
|
|
5O7G
 
 | |
5OLU
 
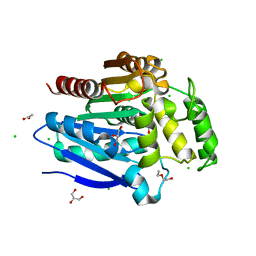 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
6TT0
 
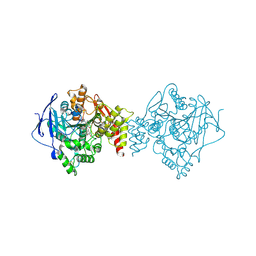 | | Crystal structure of a potent and reversible dual binding site Acetylcholinesterase chiral inhibitor | | Descriptor: | (1~{R},3~{S})-~{N}-(6,7-dimethoxy-2-oxidanylidene-chromen-3-yl)-3-[(phenylmethyl)amino]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | de la Mora, E, Mangiatordi, G.F, Belviso, B.D, Caliandro, R, Colletier, J.P, Catto, M. | | Deposit date: | 2019-12-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.80003023 Å) | | Cite: | Chiral Separation, X-ray Structure, and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
5D3Z
 
 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase in complex with a small phosphonate inhibitor | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
5D3K
 
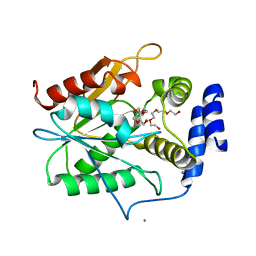 | | Crystal structure of the thioesterase domain of deoxyerythronolide B synthase | | Descriptor: | CALCIUM ION, Erythronolide synthase, modules 5 and 6, ... | | Authors: | Bergeret, F, Argyropoulos, P, Boddy, C.N, Schmeing, T.M. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards a characterization of the structural determinants of specificity in the macrocyclizing thioesterase for deoxyerythronolide B biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
5NF5
 
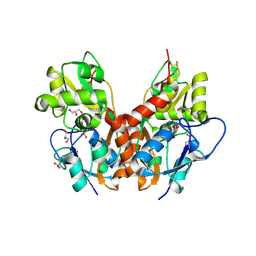 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with CIP-AS at 2.85 A resolution | | Descriptor: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | Deposit date: | 2017-03-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5O4F
 
 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with the agonist LM-12b at 2.10 A resolution | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Moellerud, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NIH
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist LM-12b at 1.3 A resolution. | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NEB
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with LM-12b at 2.05 A resolution | | Descriptor: | (3~{a}~{R},4~{S},6~{a}~{R})-1-methyl-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-c]pyrazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Moellerud, S, Frydenvang, K, Laulumaa, S, Kastrup, J.S. | | Deposit date: | 2017-03-10 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NG9
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with agonist CIP-AS at 1.15 A resolution. | | Descriptor: | (2~{S},3~{R},4~{R})-3-(carboxycarbonyl)-4-oxidanyl-pyrrolidine-2-carboxylic acid, (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, CITRATE ANION, ... | | Authors: | Laulumaa, S, Frydenvang, K.A, Winther, S, Kastrup, J.S. | | Deposit date: | 2017-03-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
5NF6
 
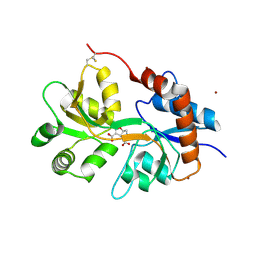 | | Structure of GluK3 ligand-binding domain (S1S2) in complex with CIP-AS at 2.55 A resolution | | Descriptor: | (3~{a}~{S},4~{S},6~{a}~{R})-4,5,6,6~{a}-tetrahydro-3~{a}~{H}-pyrrolo[3,4-d][1,2]oxazole-3,4-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Venskutonyte, R, Thorsen, T.S, Kastrup, J.S. | | Deposit date: | 2017-03-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Affinity of Two Bicyclic Glutamate Analogues at AMPA and Kainate Receptors.
ACS Chem Neurosci, 8, 2017
|
|
