5O7G
 
 | |
5OLU
 
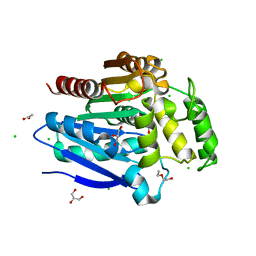 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
5N2B
 
 | |
4UT1
 
 | |
8PUO
 
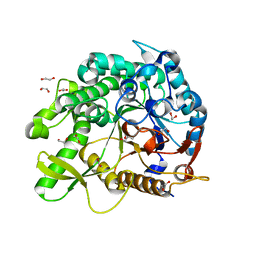 | |
2F8F
 
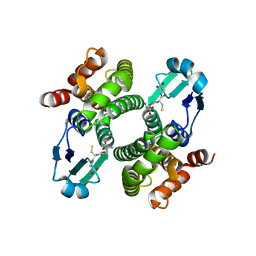 | | Crystal structure of the Y10F mutant of the gluathione s-transferase from schistosoma haematobium | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 28 kDa | | Authors: | Gourlay, L.J, Baiocco, P, Angelucci, F, Miele, A.E, Bellelli, A, Brunori, M. | | Deposit date: | 2005-12-02 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Mechanism of GSH Activation in Schistosoma haematobium Glutathione-S-transferase by Site-directed Mutagenesis and X-ray Crystallography.
J.Mol.Biol., 360, 2006
|
|
2XHZ
 
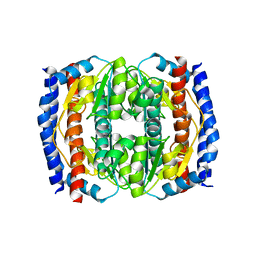 | | Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography | | Descriptor: | ARABINOSE 5-PHOSPHATE ISOMERASE | | Authors: | Gourlay, L.J, Sommaruga, S, Nardini, M, Sperandeo, P, Deho, G, Polissi, A, Bolognesi, M. | | Deposit date: | 2010-06-24 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the Active Site of the Sugar Isomerase Domain from E. Coli Arabinose-5-Phosphate Isomerase Via X-Ray Crystallography.
Protein Sci., 19, 2010
|
|
6QJ6
 
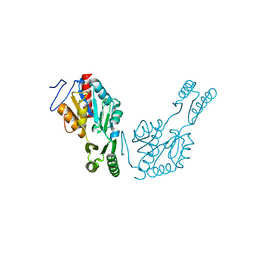 | | The structure of Trehalose-6-phosphatase from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-01-23 | | Release date: | 2020-01-15 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
6RCZ
 
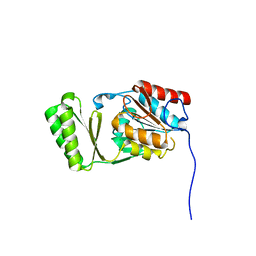 | | The structure of Burkholderia pseudomallei trehalose-6-phosphatase | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Trehalose 6-phosphate phosphatase | | Authors: | Gourlay, L.J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functional and structural analysis of trehalose-6-phosphate phosphatase from Burkholderia pseudomallei: Insights into the catalytic mechanism.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
3FAW
 
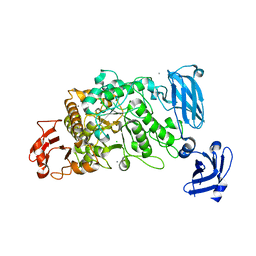 | | Crystal Structure of the Group B Streptococcus Pullulanase SAP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Reticulocyte binding protein | | Authors: | Gourlay, L.J. | | Deposit date: | 2008-11-18 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group B Streptococcus pullulanase crystal structures in the context of a novel strategy for vaccine development
J.Bacteriol., 191, 2009
|
|
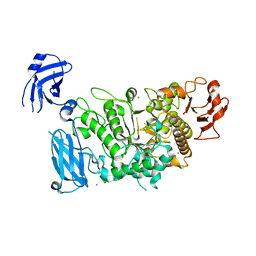
3FAX
 
 | |
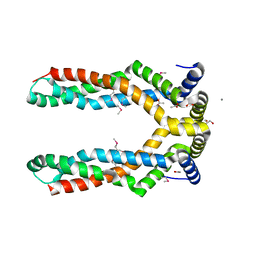
6F03
 
 | |
6FYQ
 
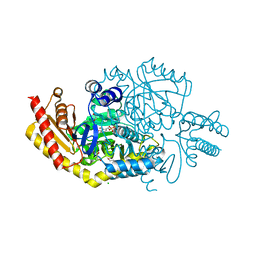 | |
6GWI
 
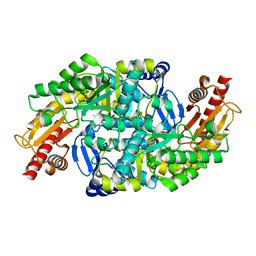 | | The crystal structure of Halomonas elongata amino-transferase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Gourlay, L.J. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-08 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Widely applicable background depletion step enables transaminase evolution through solid-phase screening.
Chem Sci, 10, 2019
|
|
6XWB
 
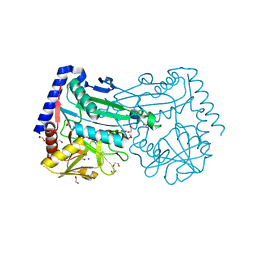 | |
6ZIV
 
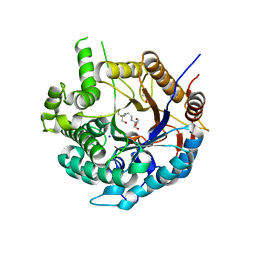 | | Crystal structure of a Beta-glucosidase from Alicyclobacillus acidiphilus | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, ... | | Authors: | Gourlay, L.J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-29 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Producing natural vanilla extract from green vanilla beans using a beta-glucosidase from Alicyclobacillus acidiphilus.
J.Biotechnol., 329, 2021
|
|
6Z42
 
 | |
2CK1
 
 | | The structure of oxidised cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin-A from Schistosoma Mansoni: Evidence for Redox- Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
2CMT
 
 | | The structure of reduced cyclophilin A from s. mansoni | | Descriptor: | ACETATE ION, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE E | | Authors: | Gourlay, L.J, Angelucci, F, Bellelli, A, Boumis, G, Miele, A.E, Brunori, M. | | Deposit date: | 2006-05-12 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of Two Redox States of Cyclophilin a from Schistosoma Mansoni. Evidence for Redox Regulation of Peptidyl-Prolyl Cis-Trans Isomerase Activity.
J.Biol.Chem., 282, 2007
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
6YN7
 
 | |
5N2C
 
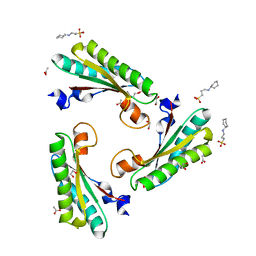 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ACETATE ION, ... | | Authors: | Matterazzo, E, Bolognesi, M, Gourlay, L.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designing Probes for Immunodiagnostics: Structural Insights into an Epitope Targeting Burkholderia Infections.
ACS Infect Dis, 3, 2017
|
|
7QQE
 
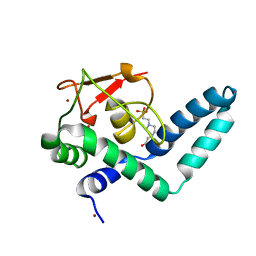 | | Nuclear factor one X - NFIX in P41212 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nuclear factor 1 X-type, ZINC ION | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
7QQD
 
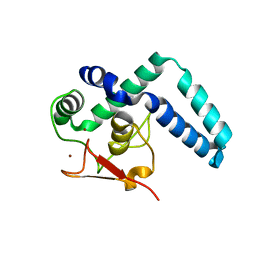 | | Nuclear factor one X - NFIX in P21 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NFI binding site (forward), NFI binding site (reverse), ... | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
4USX
 
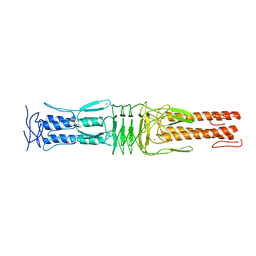 | | The Structure of the C-terminal YadA-like domain of BPSL2063 from Burkholderia pseudomallei | | Descriptor: | MAGNESIUM ION, TRIMERIC AUTOTRANSPORTER ADHESIN | | Authors: | Perletti, L, Gourlay, L.J, Peano, C, Pietrelli, A, DeBellis, G, Deantonio, C, Santoro, C, Sblattero, D, Bolognesi, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selecting Soluble/Foldable Protein Domains Through Single-Gene or Genomic Orf Filtering: Structure of the Head Domain of Burkholderia Pseudomallei Antigen Bpsl2063.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
