7EAM
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7E7E
 
 | | The co-crystal structure of ACE2 with Fab | | Descriptor: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7DA4
 
 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5VKZ
 
 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
6BG9
 
 | |
2H6M
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | N-ACETYL-LEUCYL-ALANYL-ALANYL-(N,N-DIMETHYL)-GLUTAMINE-(1,4-DIOXO-3,4-DIHYDRO-1H-PHTHALAZIN-2-YL)METHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Picornain 3C | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-05-31 | | Release date: | 2006-08-08 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors.
J.Mol.Biol., 361, 2006
|
|
2JRH
 
 | |
3RER
 
 | | Crystal structure of E. coli Hfq in complex with AU6A RNA and ADP | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*UP*A)-3', ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, W.W, Wu, J.H, Shi, Y.Y. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cooperation of Escherichia coli Hfq hexamers in DsrA binding.
Genes Dev., 25, 2011
|
|
4LMN
 
 | | Crystal Structure of MEK1 kinase bound to GDC0973 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers.
Nature, 501, 2013
|
|
2JR7
 
 | | Solution structure of human DESR1 | | Descriptor: | DPH3 homolog, ZINC ION | | Authors: | Wu, F, Wu, J, Shi, Y. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human DESR1, a CSL zinc-binding protein
Proteins, 71, 2008
|
|
3RL7
 
 | | Crystal structure of hDLG1-PDZ1 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
3RES
 
 | |
3RGQ
 
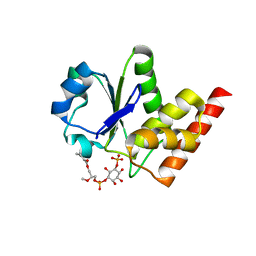 | | Crystal Structure of PTPMT1 in complex with PI(5)P | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Protein-tyrosine phosphatase mitochondrial 1 | | Authors: | Xiao, J, Engel, J.L. | | Deposit date: | 2011-04-08 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RL8
 
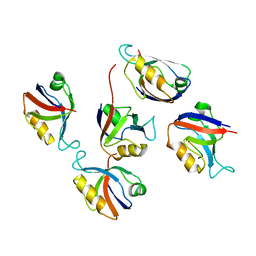 | | Crystal structure of hDLG1-PDZ2 complexed with APC | | Descriptor: | 11-mer peptide from Adenomatous polyposis coli protein, Disks large homolog 1 | | Authors: | Zhang, Z, Li, H, Wu, G. | | Deposit date: | 2011-04-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the recognition of adenomatous polyposis coli by the Discs Large 1 protein.
Plos One, 6, 2011
|
|
7E9P
 
 | |
7E9N
 
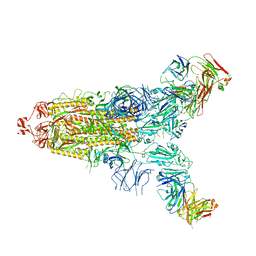 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
 | |
8SBG
 
 | |
8SIJ
 
 | | Crystal structure of F. varium tryptophanase | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Tryptophanase 1, ... | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
8SL7
 
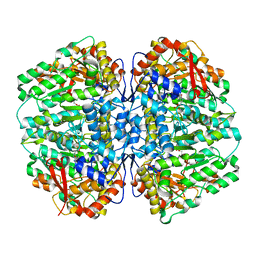 | | Butyricicoccus sp. BIOML-A1 tryptophanase complex with (3S) ALG-05 | | Descriptor: | (E)-3-[(3S)-3-chloro-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, Tryptophanase | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
5YJ8
 
 | |
3P10
 
 | |
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
