1P4Q
 
 | | Solution structure of the CITED2 transactivation domain in complex with the p300 CH1 domain | | Descriptor: | Cbp/p300-interacting transactivator 2, E1A-associated protein p300, ZINC ION | | Authors: | Freedman, S.J, Sun, Z.-Y.J, Kung, A.L, France, D.S, Wagner, G, Eck, M.J. | | Deposit date: | 2003-04-23 | | Release date: | 2003-07-01 | | Last modified: | 2017-02-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2.
Nat.Struct.Biol., 10, 2003
|
|
1OX9
 
 | | Crystal structure of SspB-ssrA complex | | Descriptor: | Stringent starvation protein B, ssrA | | Authors: | Song, H.K, Eck, M.J. | | Deposit date: | 2003-04-01 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of degradation signal recognition by SspB, a specificity-enhancing factor for the ClpXP proteolytic machine
Mol.Cell, 12, 2003
|
|
1NHL
 
 | | SNAP-23N Structure | | Descriptor: | Synaptosomal-associated protein 23 | | Authors: | Freedman, S.J, Song, H.K, Xu, Y, Eck, M.J. | | Deposit date: | 2002-12-19 | | Release date: | 2003-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homotetrameric Structure of the SNAP-23 N-terminal Coiled-coil Domain
J.Biol.Chem., 278, 2003
|
|
5TWU
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TX3
 
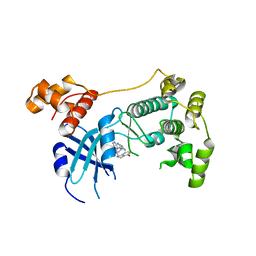 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-[(1S)-4-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-2-({3-[(pyrrolidin-1-yl)methyl]phenyl}amino)-5,7-dihydro-6H-pyrrolo[2,3-d]pyrimidin-6-one, Maternal embryonic leucine zipper kinase | | Authors: | Li, Q, Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S, Eck, M.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
1Q68
 
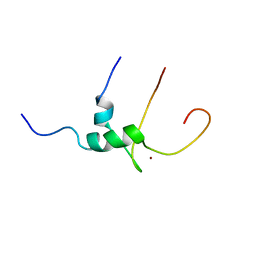 | | Solution structure of T-cell surface glycoprotein CD4 and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD4, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
5WIN
 
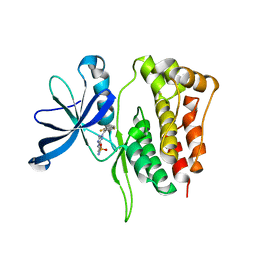 | | JAK2 Pseudokinase in complex with JNJ7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
1NTV
 
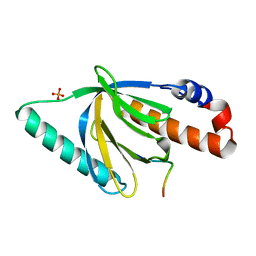 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1Q69
 
 | | Solution structure of T-cell surface glycoprotein CD8 alpha chain and Proto-oncogene tyrosine-protein kinase LCK fragments | | Descriptor: | Proto-oncogene tyrosine-protein kinase LCK, T-cell surface glycoprotein CD8 alpha chain, ZINC ION | | Authors: | Kim, P.W, Sun, Z.Y, Blacklow, S.C, Wagner, G, Eck, M.J. | | Deposit date: | 2003-08-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8.
Science, 301, 2003
|
|
1NU2
 
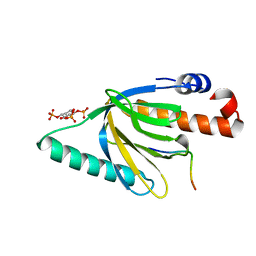 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
5WFN
 
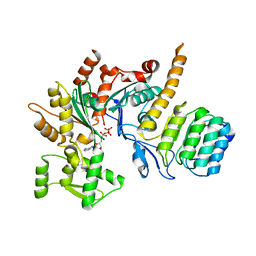 | | Revised model of leiomodin 2-mediated actin regulation (alternate refinement of PDB 4RWT) | | Descriptor: | Actin-5C, Leiomodin-2, MAGNESIUM ION, ... | | Authors: | Yurtsever, Z, Eck, M.J, Dominguez, R. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of leiomodin 2 in complex with actin: a structural and functional reexamination
Biophys.J., 113, 2017
|
|
5WIJ
 
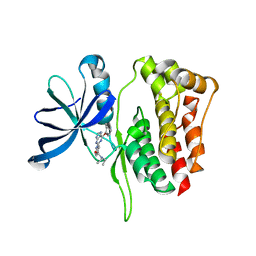 | | JAK2 Pseudokinase in complex with NU6140 | | Descriptor: | 4-{[6-(cyclohexylmethoxy)-7H-purin-2-yl]amino}-N,N-diethylbenzamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIL
 
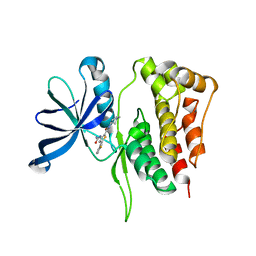 | | JAK2 Pseudokinase in complex with AZD7762 | | Descriptor: | 5-(3-fluorophenyl)-N-[(3S)-3-piperidyl]-3-ureido-thiophene-2-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIK
 
 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
6V5N
 
 | | EGFR(T790M/V948R) in complex with LN2084 | | Descriptor: | 3-[4-(4-fluorophenyl)-5-(2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)-1H-imidazol-2-yl]propan-1-ol, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V6O
 
 | | EGFR(T790M/V948R) in complex with LN2380 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(3-hydroxypropyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VHP
 
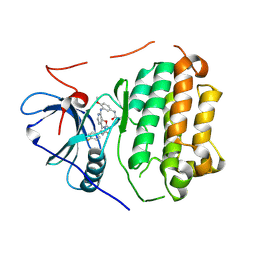 | | Wild type EGFR in complex with LN2899 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(4-{4-(4-fluorophenyl)-2-[(2-methoxyethyl)sulfanyl]-1H-imidazol-5-yl}pyridin-2-yl)amino]-4-methoxyphenyl}propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V2U
 
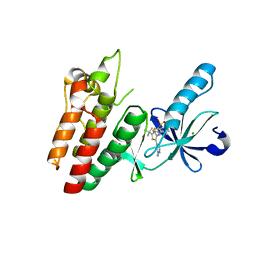 | |
6V6K
 
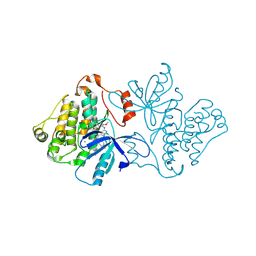 | | EGFR(T790M/V948R) in complex with LN2057 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(methylsulfanyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V2W
 
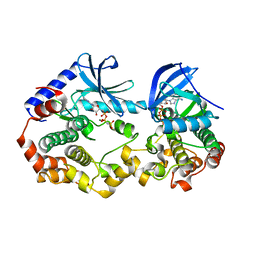 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Park, E, Eck, M.J. | | Deposit date: | 2019-11-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6V34
 
 | |
6VH4
 
 | | Wild type EGFR in complex with LN2380 | | Descriptor: | Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(3-hydroxypropyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V66
 
 | | EGFR(T790M/V948R) in complex with LN2899 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Epidermal growth factor receptor, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V5P
 
 | | EGFR(T790M/V948R) in complex with LN2725 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-(3-methoxypropyl)-1H-imidazol-5-yl]-2-phenyl-3H-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
