6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
1CW2
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLSULFINYL)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-25 | | Release date: | 1999-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
5LAX
 
 | | Crystal structure of HLA_DRB1*04:01 in complex with alpha-enolase peptide 26-40 | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-4 beta chain, ... | | Authors: | Dubnovitsky, A, Kozhukh, G, Sandalova, T, Achour, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Characterization of a Novel HLA-DRB1*04:01-Restricted alpha-Enolase T Cell Epitope in Rheumatoid Arthritis.
Front Immunol, 7, 2016
|
|
1CHL
 
 | | NMR SEQUENTIAL ASSIGNMENTS AND SOLUTION STRUCTURE OF CHLOROTOXIN, A SMALL SCORPION TOXIN THAT BLOCKS CHLORIDE CHANNELS | | Descriptor: | CHLOROTOXIN | | Authors: | Lippens, G, Najib, J, Wodak, S.J, Tartar, A. | | Deposit date: | 1994-11-09 | | Release date: | 1995-02-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels.
Biochemistry, 34, 1995
|
|
4P5D
 
 | | CRYSTAL STRUCTURE OF RAT DNPH1 (RCL) WITH 6-NAPHTHYL-PURINE-RIBOSIDE-MONOPHOSPHATE | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 6-(naphthalen-2-yl)-9-(5-O-phosphono-beta-D-ribofuranosyl)-9H-purine, SULFATE ION | | Authors: | Padilla, A, Labesse, G, Kaminski, P.A. | | Deposit date: | 2014-03-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 6-(Hetero)Arylpurine nucleotides as inhibitors of the oncogenic target DNPH1: Synthesis, structural studies and cytotoxic activities.
Eur.J.Med.Chem., 85C, 2014
|
|
2VUV
 
 | | Crystal structure of Codakine at 1.3A resolution | | Descriptor: | CALCIUM ION, CITRIC ACID, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
2W4S
 
 | | novel RNA-binding domain in Cryptosporidium parvum at 2.5 angstrom resolution | | Descriptor: | Ankyrin-repeat protein | | Authors: | Varrot, A, Mackereth, C, Mourao, A, Sattler, M, Cusack, S. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and RNA Recognition by the Snrna and Snorna Transport Factor Phax.
RNA, 16, 2010
|
|
2WAM
 
 | | Crystal structure of Mycobacterium tuberculosis unknown function protein Rv2714 | | Descriptor: | CONSERVED HYPOTHETICAL ALANINE AND LEUCINE RICH PROTEIN, GLYCEROL | | Authors: | Bellinzoni, M, Grana, M, Buschiazzo, A, Miras, I, Haouz, A, Alzari, P.M. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium Tuberculosis Rv2714, a Representative of a Duplicated Gene Family in Actinobacteria.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4PSG
 
 | |
6NSQ
 
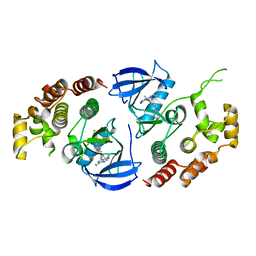 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
7QBU
 
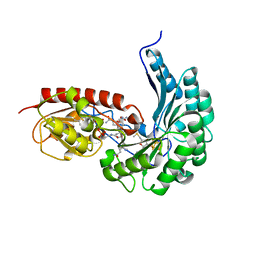 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7F8R
 
 | |
1Q0Q
 
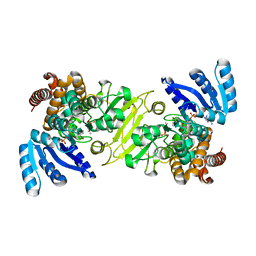 | | Crystal structure of DXR in complex with the substrate 1-deoxy-D-xylulose-5-phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mac Sweeney, A, Lange, R, D'Arcy, A, Douangamath, A, Surivet, J.-P, Oefner, C. | | Deposit date: | 2003-07-17 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of E.coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation.
J.Mol.Biol., 345, 2005
|
|
7QBT
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, FE (III) ION, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
8PH6
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K2[Ru2(DPhF)(CO3)3] in condition B | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme C, Ru2(DPhF)(CO3)2(Formate), ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
4XRT
 
 | | Crystal structure of the di-domain ARO/CYC StfQ from the steffimycin biosynthetic pathway | | Descriptor: | FORMIC ACID, StfQ Aromatase/Cyclase | | Authors: | Tsai, S.C, Caldara-Festin, G.M, Jackson, D.R, Aguilar, S, Patel, A, Nguyen, M, Sasaki, E, Valentic, T.R, Barajas, J.F, Vo, M, Khanna, A, Liu, H.-W. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural and functional analysis of two di-domain aromatase/cyclases from type II polyketide synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1D59
 
 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1992-02-25 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
8PH8
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)2(O2CCH3)2] in condition A | | Descriptor: | 1-oxidanyl-2,4,6,8-tetraphenyl-2,4,6,8-tetraza-1$l^{4},5$l^{3}-diruthenabicyclo[3.3.0]octane, Lysozyme C, SODIUM ION, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
6X5N
 
 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
7QBV
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
1D8X
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*TP*GP*AP*GP*G)-3', COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-26 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
8PH7
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DPhF)(O2CCH3)3] in condition A | | Descriptor: | Lysozyme C, SODIUM ION, SUCCINIC ACID, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-19 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | Steric hindrance and charge influence on the cytotoxic activity and protein binding properties of diruthenium complexes.
Int.J.Biol.Macromol., 253, 2023
|
|
1D98
 
 | | THE STRUCTURE OF AN OLIGO(DA).OLIGO(DT) TRACT AND ITS BIOLOGICAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Nelson, H.C.M, Finch, J.T, Luisi, B.F, Klug, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of an oligo(dA).oligo(dT) tract and its biological implications.
Nature, 330, 1987
|
|
1D9I
 
 | | STRUCTURE OF THROMBIN COMPLEXED WITH SELECTIVE NON-ELECTOPHILIC INHIBITORS HAVING CYCLOHEXYL MOIETIES AT P1 | | Descriptor: | (5S)-N-[(trans-4-aminocyclohexyl)methyl]-1,3-dioxo-2-[2-(phenylsulfonyl)ethyl]-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazine-5-carboxamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R, Tulinsky, A. | | Deposit date: | 1999-10-27 | | Release date: | 2000-10-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
