1CHL
 
 | | NMR SEQUENTIAL ASSIGNMENTS AND SOLUTION STRUCTURE OF CHLOROTOXIN, A SMALL SCORPION TOXIN THAT BLOCKS CHLORIDE CHANNELS | | Descriptor: | CHLOROTOXIN | | Authors: | Lippens, G, Najib, J, Wodak, S.J, Tartar, A. | | Deposit date: | 1994-11-09 | | Release date: | 1995-02-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR sequential assignments and solution structure of chlorotoxin, a small scorpion toxin that blocks chloride channels.
Biochemistry, 34, 1995
|
|
1GHC
 
 | | HOMO-AND HETERONUCLEAR TWO-DIMENSIONAL NMR STUDIES OF THE GLOBULAR DOMAIN OF HISTONE H1: FULL ASSIGNMENT, TERTIARY STRUCTURE, AND COMPARISON WITH THE GLOBULAR DOMAIN OF HISTONE H5 | | Descriptor: | GH1 | | Authors: | Cerf, C, Lippens, G, Ramakrishnan, V, Muyldermans, S, Segers, A, Wyns, L, Wodak, S.J, Hallenga, K. | | Deposit date: | 1994-05-16 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: full assignment, tertiary structure, and comparison with the globular domain of histone H5.
Biochemistry, 33, 1994
|
|
1TXK
 
 | | Crystal structure of Escherichia coli OpgG | | Descriptor: | Glucans biosynthesis protein G, SODIUM ION | | Authors: | Hanoulle, X, Rollet, E, Clantin, B, Landrieu, I, Odberg-Ferragut, C, Lippens, G, Bohin, J.P, Villeret, V. | | Deposit date: | 2004-07-05 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Escherichia coli OpgG, a Protein Required for the Biosynthesis of Osmoregulated Periplasmic Glucans.
J.Mol.Biol., 342, 2004
|
|
6HTV
 
 | | Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with isomaltotetraose | | Descriptor: | Alternansucrase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Claverie, M, Cioci, G, Remaud-Simeon, M, Moulis, C, Lippens, G. | | Deposit date: | 2018-10-04 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Futile Encounter Engineering of the DSR-M Dextransucrase Modifies the Resulting Polymer Length.
Biochemistry, 58, 2019
|
|
1J6Y
 
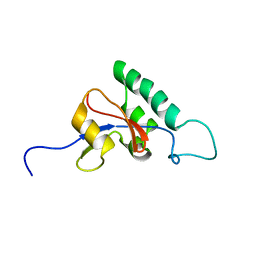 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
1T3K
 
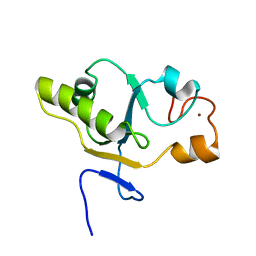 | | NMR structure of a CDC25-like dual-specificity tyrosine phosphatase of Arabidopsis thaliana | | Descriptor: | Dual-specificity tyrosine phosphatase, ZINC ION | | Authors: | Landrieu, I, da Costa, M, De Veylder, L, Dewitte, F, Vandepoele, K, Hassan, S, Wieruszeski, J.M, Faure, J.D, Inze, D, Lippens, G. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-07 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | A small CDC25 dual-specificity tyrosine-phosphatase isoform in Arabidopsis thaliana.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6HT4
 
 | |
2M5L
 
 | |
3TP0
 
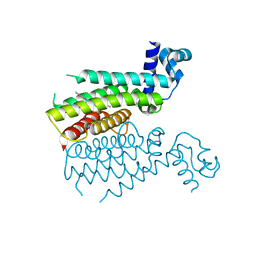 | | Structural activation of the transcriptional repressor EthR from M. tuberculosis by single amino-acid change mimicking natural and synthetic ligands | | Descriptor: | 3-oxo-3-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}propanenitrile, HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Blondiaux, N, Willery, E, Hoos, S, Lecat-Guillet, N, Lens, Z, Wohlkonig, A, Wintjens, R, Soror, S, Fr nois, F, Diri, B, Villeret, V, England, P, Lippens, G, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2011
|
|
2REL
 
 | | SOLUTION STRUCTURE OF R-ELAFIN, A SPECIFIC INHIBITOR OF ELASTASE, NMR, 11 STRUCTURES | | Descriptor: | R-ELAFIN | | Authors: | Francart, C, Dauchez, M, Alix, A.J.P, Lippens, G. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of R-elafin, a specific inhibitor of elastase.
J.Mol.Biol., 268, 1997
|
|
1HTX
 
 | | SOLUTION STRUCTURE OF THE MAIN ALPHA-AMYLASE INHIBITOR FROM AMARANTH SEEDS | | Descriptor: | ALPHA-AMYLASE INHIBITOR AAI | | Authors: | Martins, J.C, Enassar, M, Willem, R, Wieruzeski, J.M, Lippens, G, Wodak, S.J. | | Deposit date: | 2001-01-02 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the main alpha-amylase inhibitor from amaranth seeds.
Eur.J.Biochem., 268, 2001
|
|
1I8G
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH CDC25 PHOSPHOTHREONINE PEPTIDE | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 3, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1I8H
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN COMPLEXED WITH HUMAN TAU PHOSPHOTHREONINE PEPTIDE | | Descriptor: | MICROTUBULE-ASSOCIATED PROTEIN TAU, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-14 | | Release date: | 2001-07-18 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
1I6C
 
 | | SOLUTION STRUCTURE OF PIN1 WW DOMAIN | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Wintjens, R, Wieruszeski, J.-M, Drobecq, H, Lippens, G, Landrieu, I. | | Deposit date: | 2001-03-02 | | Release date: | 2001-07-18 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides.
J.Biol.Chem., 276, 2001
|
|
7NEK
 
 | | SusD protein from human gut uncultured Bacteroides | | Descriptor: | GLYCEROL, RagB SusD domain containing protein | | Authors: | Cioci, C, Tauzin, A, Wang, Z, Machado, B, Lippens, G, Potocki-Veronese, G. | | Deposit date: | 2021-02-04 | | Release date: | 2022-03-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SusD protein from human gut uncultured Bacteroides
To Be Published
|
|
