8OIF
 
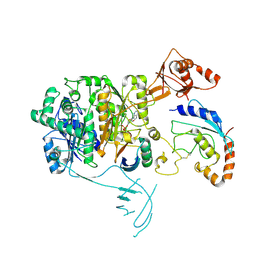 | | Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Wallace, I, Kheewoong, B, Prabu, J.R, Vollrath, R, von Gronau, S, Schulman, B.A, Swatek, K.N. | | Deposit date: | 2023-03-22 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Insights into the ISG15 transfer cascade by the UBE1L activating enzyme.
Nat Commun, 14, 2023
|
|
8PMQ
 
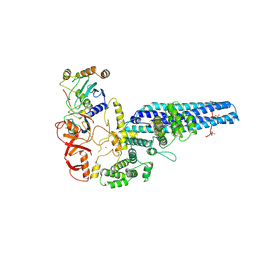 | | Catalytic module of yeast GID E3 ligase bound to multiphosphorylated Ubc8~ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PJN
 
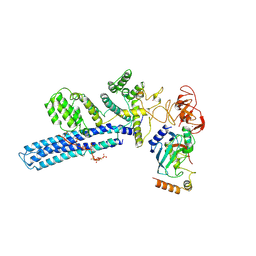 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | Descriptor: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | Deposit date: | 2023-06-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PDA
 
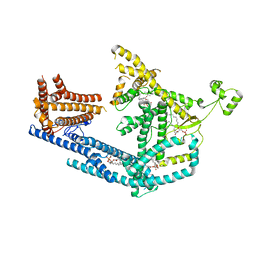 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PD0
 
 | | cryo-EM structure of Doa10 in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | Descriptor: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
4CBC
 
 | |
8BD0
 
 | |
8CQF
 
 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis Complexed with Rearranged Acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Skagseth, S, Griese, J.J, Lund, B.A, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
8CQG
 
 | | Crystal Structure of a Chimeric Alpha-Amylase from Pseudoalteromonas Haloplanktis | | Descriptor: | 1,2-ETHANEDIOL, Alpha-amylase, CALCIUM ION, ... | | Authors: | Skagseth, S, Lund, B.A, Griese, J.J, van der Ent, F, Aqvist, J. | | Deposit date: | 2023-03-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computational design of the temperature optimum of an enzyme reaction.
Sci Adv, 9, 2023
|
|
8CDK
 
 | |
8CDJ
 
 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
8C07
 
 | |
8C06
 
 | |
2RTB
 
 | | APOSTREPTAVIDIN, PH 3.32, SPACE GROUP I222 | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTR
 
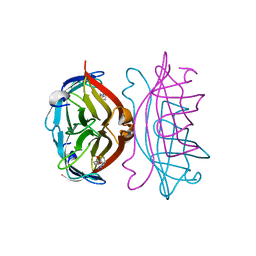 | | STREPTAVIDIN-2-IMINOBIOTIN COMPLEX, PH 4.0, SPACE GROUP I222 | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTQ
 
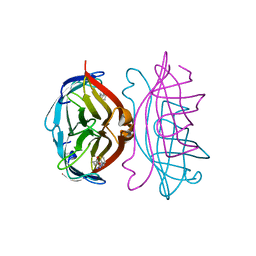 | |
2RTP
 
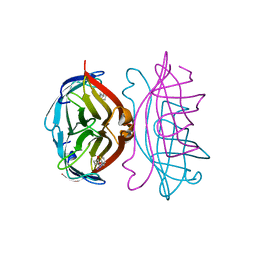 | |
2RTN
 
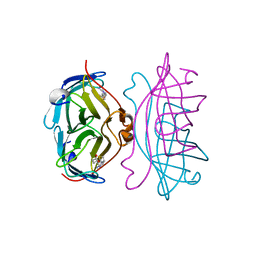 | | STREPTAVIDIN-2-IMINOBIOTIN COMPLEX, PH 2.0, SPACE GROUP I222 | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTI
 
 | | STREPTAVIDIN-GLYCOLURIL COMPLEX, PH 2.50, SPACE GROUP I222 | | Descriptor: | FORMIC ACID, GLYCOLURIL, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTO
 
 | | STREPTAVIDIN-2-IMINOBIOTIN COMPLEX, PH 2.6, SPACE GROUP I222 | | Descriptor: | 2-IMINOBIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2RTM
 
 | |
2RTA
 
 | | APOSTREPTAVIDIN, PH 2.97, SPACE GROUP I4122 | | Descriptor: | STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-09-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
