5ZQ0
 
 | | Crystal structure of spRlmCD with U747loop RNA | | Descriptor: | RNA (5'-R(*GP*UP*(MUM)P*GP*AP*AP*AP*A)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
5ZQ8
 
 | | Crystal structure of spRlmCD with U747 stemloop RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(*CP*CP*GP*UP*(MUM)P*GP*AP*AP*AP*AP*GP*G)-3'), S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
5ZTH
 
 | | Crystal structure of spRlmCD with U1939loop RNA at 3.24 angstrom | | Descriptor: | RNA (5'-R(P*AP*AP*AP*(MUM)P*UP*CP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
5T18
 
 | | Crystal structure of Bruton agammabulinemia tyrosine kinase complexed with BMS-986142 aka (2s)-6-fluoro-5-[3-(8-fluoro-1-methyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-3-yl)-2-methylphenyl]-2-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1h-carbazole-8-carboxamide | | Descriptor: | 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-08-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton's Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers.
J. Med. Chem., 59, 2016
|
|
2GJJ
 
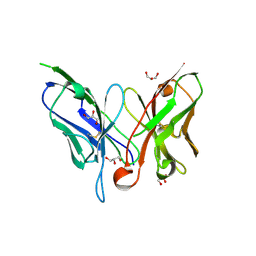 | | Crystal structure of a single chain antibody scA21 against Her2/ErbB2 | | Descriptor: | A21 single-chain antibody fragment against erbB2, GLYCEROL | | Authors: | Zhu, Z. | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope mapping and structural analysis of an anti-ErbB2 antibody A21: Molecular basis for tumor inhibitory mechanism
Proteins, 70, 2007
|
|
5IHL
 
 | |
3QWR
 
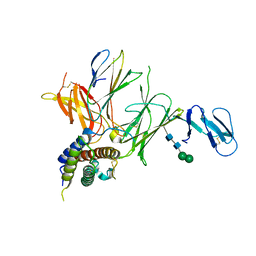 | | Crystal structure of IL-23 in complex with an adnectin | | Descriptor: | ADNECTIN, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Wei, A, Sheriff, S. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of adnectin/protein complexes reveal an expanded binding footprint.
Structure, 20, 2012
|
|
3QWQ
 
 | |
5XHS
 
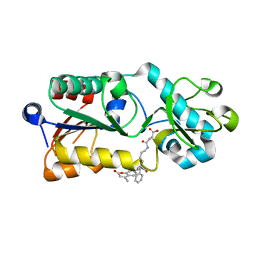 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
6J71
 
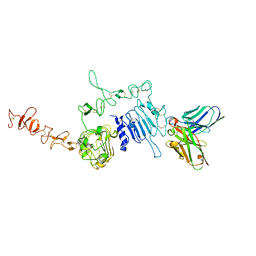 | | HuA21-scFv in complex with the extracellular domain(ECD) of HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-HER2 humanized antibody HuA21, beta-D-mannopyranose, ... | | Authors: | Wang, Z, Guo, G, Cheng, B, Zhu, Z, Niu, L, Zhang, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural insight into a matured humanized monoclonal antibody HuA21 against HER2-overexpressing cancer cells.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5YYZ
 
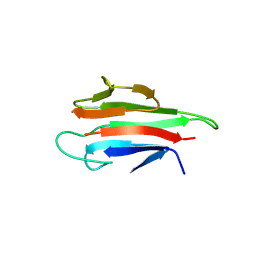 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | Descriptor: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YYX
 
 | | Crystal Structure of the MEK1 FHA domain | | Descriptor: | Meiosis-specific serine/threonine-protein kinase MEK1 | | Authors: | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4XHQ
 
 | |
4X9H
 
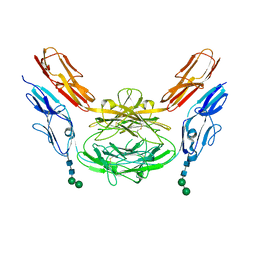 | | Crystal structure of Dscam1 isoform 8.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform AP, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
5TKB
 
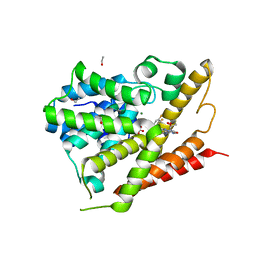 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 4D IN COMPLEX WITH A TETRAFLUORANLINE COMPOUND | | Descriptor: | ETHANOL, MAGNESIUM ION, N-[(2R)-2,3-dihydroxy-2-methylpropyl]-8-(methylamino)-6-[(2,3,5,6-tetrafluorophenyl)amino]imidazo[1,2-b]pyridazine-3-carboxamide, ... | | Authors: | Sack, J.S. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
5TKD
 
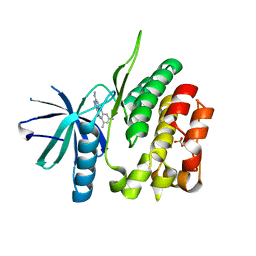 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH 6-[(3,5-DIMETHYLPHE NYL)AMINO]-8- (METHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBO XAMIDE | | Descriptor: | 6-[(3,5-dimethylphenyl)amino]-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-10-06 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of imidazo[1,2-b]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling.
Medchemcomm, 8, 2017
|
|
6FMC
 
 | | Neuropilin1-b1 domain in complex with EG01377, 0.9 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | Authors: | Yelland, T, Djordjevic, S, Fotinou, K, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6FMF
 
 | | Neuropilin-1 b1 domain in complex with EG01377; 2.8 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1, trifluoroacetic acid | | Authors: | Yelland, T, Djordjevic, S, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
5GON
 
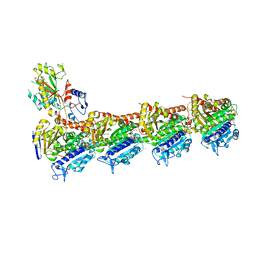 | | Structures of a beta-lactam bridged analogue in complex with tubulin | | Descriptor: | (3R,4R)-4-(4-methoxy-3-oxidanyl-phenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhou, L, Liu, Y, Cheng, L, Wang, Y. | | Deposit date: | 2016-07-28 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Potent Antitumor Activities and Structure Basis of the Chiral beta-Lactam Bridged Analogue of Combretastatin A-4 Binding to Tubulin.
J. Med. Chem., 59, 2016
|
|
8K37
 
 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K38
 
 | | The structure of bacteriophage lambda portal-adaptor | | Descriptor: | Head completion protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K36
 
 | |
8K35
 
 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
1KV6
 
 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
