2HE4
 
 | | The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | | Authors: | Papagrigoriou, E, Elkins, J.M, Berridge, G, Gileady, O, Colebrook, S, Gileadi, C, Salah, E, Savitsky, P, Pantic, N, Gorrec, F, Bunkoczi, G, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
4WSQ
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
8CIS
 
 | | The FERM domain of human moesin with two bound peptides identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C3P, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIU
 
 | | The FERM domain of human moesin mutant H288A | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIT
 
 | | The FERM domain of human moesin mutant L281R | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIR
 
 | | The FERM domain of human moesin with a bound peptide identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
2VPJ
 
 | | Crystal structure of the Kelch domain of human KLHL12 | | Descriptor: | ACETATE ION, KELCH-LIKE PROTEIN 12 | | Authors: | Keates, T, Pike, A.C.W, Bullock, A.N, Salah, E, Filippakopoulos, P, Roos, A.K, von Delft, F, Savitsky, P, Weigelt, J, Edwards, A, Arrowsmith, C.H, Bountra, C, Knapp, S. | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
3IR3
 
 | | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2) | | Descriptor: | 3-hydroxyacyl-thioester dehydratase 2, MALONATE ION | | Authors: | Ugochukwu, E, Cocking, R, Burgess-Brown, N, Pilka, E, Muniz, J, Krojer, T, Chaikuad, A, Gileadi, O, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of human 3-hydroxyacyl-thioester dehydratase 2 (HTD2)
To be Published
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
3Q4U
 
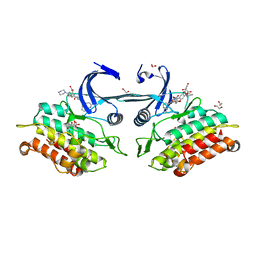 | | Crystal structure of the ACVR1 kinase domain in complex with LDN-193189 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C.D.O, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Gileadi, O, Fedorov, O, Allerston, C.K, Krojer, T, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
2OS2
 
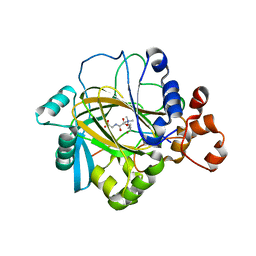 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys36 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OT7
 
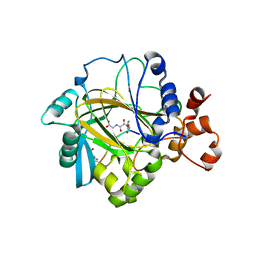 | | Crystal structure of JMJD2A complexed with histone H3 peptide monomethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-07 | | Release date: | 2007-03-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OQ6
 
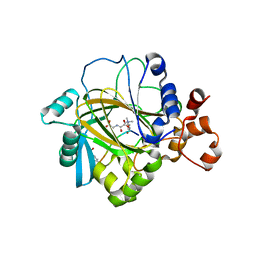 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OQ7
 
 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OX0
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide dimethylated at Lys9 | | Descriptor: | CHLORIDE ION, JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2XN4
 
 | | Crystal structure of the kelch domain of human KLHL2 (Mayven) | | Descriptor: | 1,2-ETHANEDIOL, KELCH-LIKE PROTEIN 2, SODIUM ION, ... | | Authors: | Canning, P, Hozjan, V, Cooper, C.D.O, Ayinampudi, V, Vollmar, M, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2010-07-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
2CLQ
 
 | | Structure of mitogen-activated protein kinase kinase kinase 5 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, STAUROSPORINE | | Authors: | Bunkoczi, G, Salah, E, Fedorov, O, Pike, A, Gileadi, O, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of the Human Protein Kinase Ask1.
Structure, 15, 2007
|
|
5L4Q
 
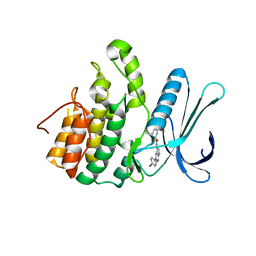 | | Crystal Structure of Adaptor Protein 2 Associated Kinase 1 (AAK1) in Complex with LKB1 (AAK1 Dual Inhibitor) | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, ~{N}-[5-(4-cyanophenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]pyridine-3-carboxamide | | Authors: | Sorrell, F.J, Williams, E, Fox, N, Abdul Azeez, K.R, Gileadi, O, von Delft, F, Edwards, A.M, Bountra, C, Elkins, J.M, Knapp, S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and Structure-Activity Relationships of 3,5-Disubstituted-pyrrolo[2,3- b]pyridines as Inhibitors of Adaptor-Associated Kinase 1 with Antiviral Activity.
J.Med.Chem., 2019
|
|
5LY1
 
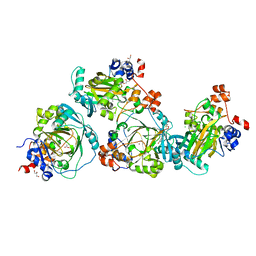 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | Descriptor: | CHLORIDE ION, CP2, GLYCEROL, ... | | Authors: | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | Deposit date: | 2016-09-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
7QGI
 
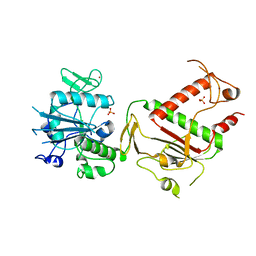 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
7QIF
 
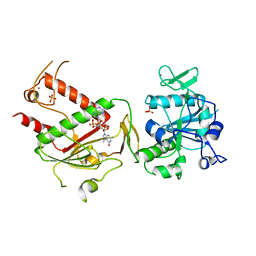 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
2C30
 
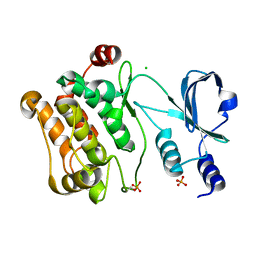 | | Crystal Structure Of The Human P21-Activated Kinase 6 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, SERINE/THREONINE-PROTEIN KINASE PAK 6 | | Authors: | Filippakopoulos, P, Berridge, G, Bray, J, Burgess, N, Colebrook, S, Das, S, Eswaran, J, Gileadi, O, Papagrigoriou, E, Savitsky, P, Smee, C, Turnbull, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Knapp, S. | | Deposit date: | 2005-10-02 | | Release date: | 2006-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the P21-Activated Kinases Pak4, Pak5, and Pak6 Reveal Catalytic Domain Plasticity of Active Group II Paks.
Structure, 15, 2007
|
|
4BZ3
 
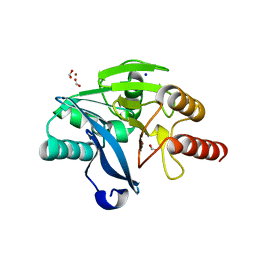 | | Crystal structure of the metallo-beta-lactamase VIM-2 | | Descriptor: | BETA-LACTAMASE VIM-2, FORMIC ACID, SODIUM ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.294 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1E
 
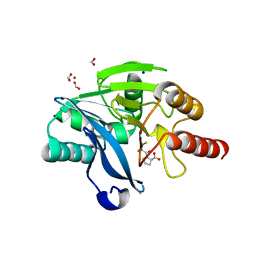 | | Crystal structure of the metallo-beta-lactamase VIM-2 with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE CLASS B VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
4C1H
 
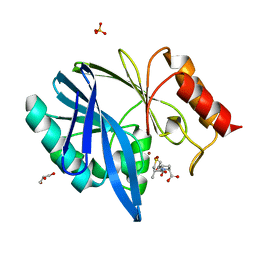 | | Crystal structure of the metallo-beta-lactamase BCII with L-captopril | | Descriptor: | BETA-LACTAMASE 2, GLYCEROL, L-CAPTOPRIL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
