8F6D
 
 | |
3DF0
 
 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4K95
 
 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
3EC3
 
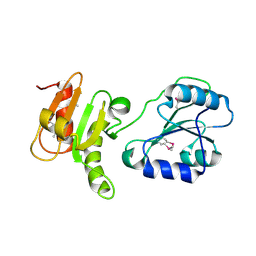 | | Crystal structure of the bb fragment of ERp72 | | Descriptor: | Protein disulfide-isomerase A4 | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2008-08-28 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of the Noncatalytic Domains and Global Fold of the Protein Disulfide Isomerase ERp72.
Structure, 17, 2009
|
|
6WUS
 
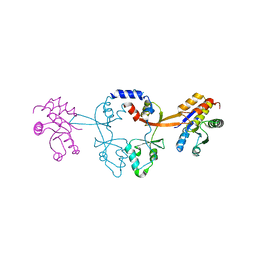 | |
6B3Y
 
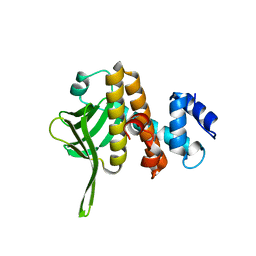 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
7SOT
 
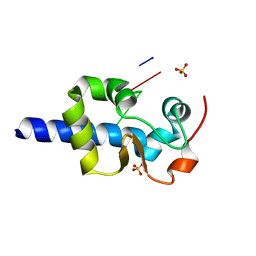 | |
7SOQ
 
 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOV
 
 | |
7SOO
 
 | | LaM domain of human LARP1 | | Descriptor: | Isoform 2 of La-related protein 1, SODIUM ION, SULFATE ION | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOP
 
 | |
6DJ3
 
 | |
7SOR
 
 | | LaM domain of human LARP1 in complex with AAA RNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Isoform 2 of La-related protein 1, ... | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
6DJW
 
 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7SOS
 
 | | LaM domain of human LARP1 in complex with AAAA RNA | | Descriptor: | Isoform 2 of La-related protein 1, POTASSIUM ION, RNA (5'-R(*AP*AP*AP*A)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis of 3'-end poly(A) RNA recognition by LARP1.
Nucleic Acids Res., 50, 2022
|
|
7SOU
 
 | |
7SOW
 
 | | LaM domain of human LARP1 in complex with UUUUUU | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of La-related protein 1, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2021-11-01 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Enhanced binding of guanylated poly(A) RNA by the LaM domain of LARP1.
Rna Biol., 21, 2024
|
|
6DJX
 
 | | Crystal Structure of pParkin-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DFD
 
 | |
2QHO
 
 | |
4IOT
 
 | | High-resolution Structure of Triosephosphate isomerase from E. coli | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Vinaik, R, Kozlov, G, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2013-01-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Triosephosphate isomerase is a common crystallization contaminant of soluble His-tagged proteins produced in Escherichia coli.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1JGN
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
4JU5
 
 | |
