7NH7
 
 | |
7Q3B
 
 | | Crystal structure of human STING in complex with 3'3'-c-(2'F,2'dA-isonucA)MP | | Descriptor: | 3'3'-c-(2'F,2'dA-isonucA)MP, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55733919 Å) | | Cite: | Discovery of isonucleotidic CDNs as potent STING agonists with immunomodulatory potential.
Structure, 30, 2022
|
|
4WAE
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.318 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
4WAG
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with MI103 inhibitor | | Descriptor: | 6-chloro-3-(3,4-dimethoxyphenyl)-2-methylimidazo[1,2-b]pyridazin-8-amine, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.407 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
6QWT
 
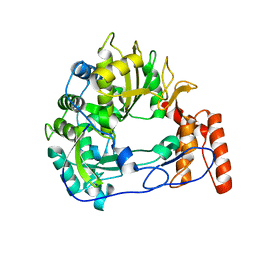 | | Sicinivirus 3Dpol RNA dependent RNA polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
6R1I
 
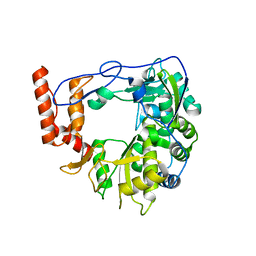 | | Structure of porcine Aichi virus polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
4YC4
 
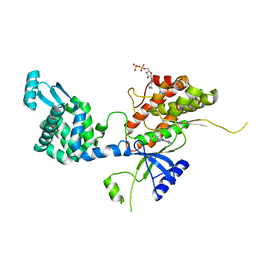 | | Crystal structure of phosphatidyl inositol 4-kinase II alpha in complex with nucleotide analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha, [(1S,3S,4S)-3-(6-amino-9H-purin-9-yl)bicyclo[2.2.1]hept-1-yl]methanol | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2015-02-19 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6YZ1
 
 | | The crystal structure of SARS-CoV-2 nsp10-nsp16 methyltransferase complex with Sinefungin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SINEFUNGIN, ZINC ION, ... | | Authors: | Krafcikova, P, Silhan, J, Nencka, R, Boura, E. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin.
Nat Commun, 11, 2020
|
|
7OB3
 
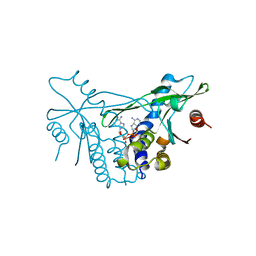 | | hSTING in complex with 3',3'-c-di-araAMP | | Descriptor: | 3',3'-c-di-araAMP, Stimulator of interferon genes protein | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2021-04-20 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Synthesis of 3'-5', 3'-5' Cyclic Dinucleotides, Their Binding Properties to the Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations
Biochemistry, 2021
|
|
7R1U
 
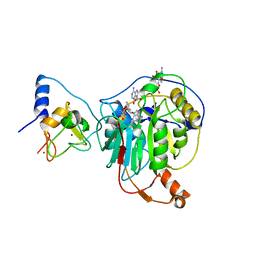 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1T
 
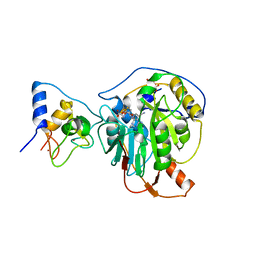 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7Q85
 
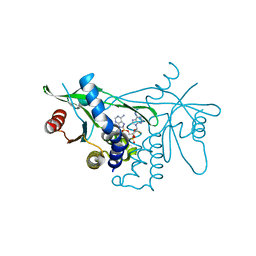 | | Crystal structure of human STING in complex with MD1193 | | Descriptor: | 9-[(1R,6R,8R,13E,15R,17R,18R)-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,16-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadec-13-en-8-yl]purin-6-amine, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Vinylphosphonate-based cyclic dinucleotides enhance STING-mediated cancer immunotherapy.
Eur.J.Med.Chem., 259, 2023
|
|
8CEV
 
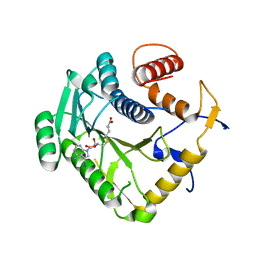 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO1119 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-iodanyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CER
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO494 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-naphthalen-1-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CEQ
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO427 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-phenylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CES
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO500 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-[2-(1~{H}-benzimidazol-2-yl)ethynyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CET
 
 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO507 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CGB
 
 | | Monkeypox virus VP39 in complex with SAH | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Silhan, J, Klima, M, Skvara, P, Boura, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8C9K
 
 | |
4WTV
 
 | | Crystal structure of the phosphatidylinositol 4-kinase IIbeta | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-beta,Endolysin,Phosphatidylinositol 4-kinase type 2-beta | | Authors: | Klima, M, Baumlova, A, Chalupska, D, Boura, E. | | Deposit date: | 2014-10-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5EUT
 
 | |
5FBW
 
 | | PI4KB in complex with Rab11 and the MI369 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | PI4KB in complex with Rab11 and the MI369 Inhibitor
To Be Published
|
|
5MRK
 
 | |
6Q69
 
 | |
6Q68
 
 | |
