5FEC
 
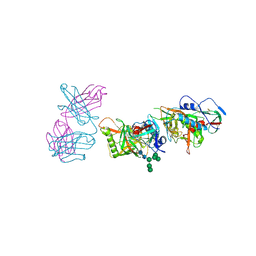 | |
5IGX
 
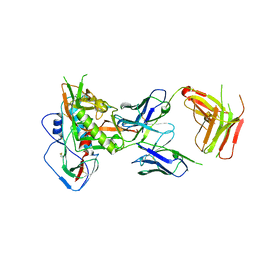 | |
5I9Q
 
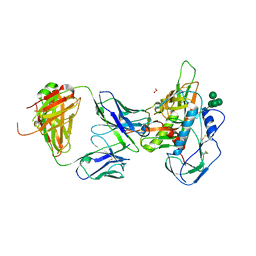 | | Crystal structure of 3BNC55 Fab in complex with 426c.TM4deltaV1-3 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC55 Fab heavy chain, ... | | Authors: | Scharf, L, Chen, C, Bjorkman, P.J. | | Deposit date: | 2016-02-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
7RKV
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKS
 
 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
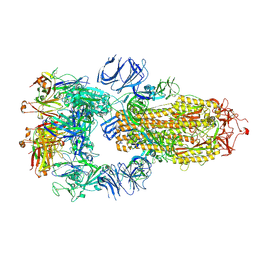
7RKU
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RFB
 
 | | Crystal structure of broadly neutralizing antibody mAb1198 in complex with Hepatitis C virus envelope glycoprotein E2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization.
Immunity, 55, 2022
|
|
7RFC
 
 | |
6MEE
 
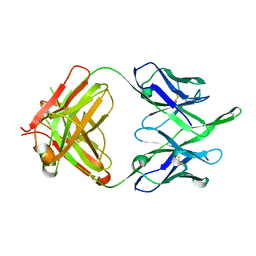 | | Crystal structure of broadly neutralizing antibody HEPC74 | | Descriptor: | antibody HEPC74 Heavy Chain, antibody HEPC74 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
6MEH
 
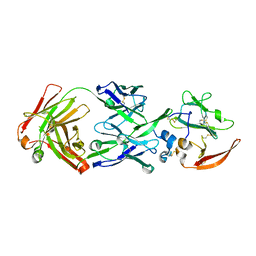 | |
6MEJ
 
 | |
6MEK
 
 | |
6MEF
 
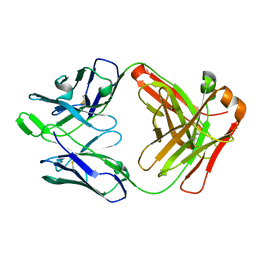 | | Crystal structure of broadly neutralizing antibody AR3C | | Descriptor: | antibody AR3C Heavy Chain, antibody AR3C Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
6MEI
 
 | |
6MED
 
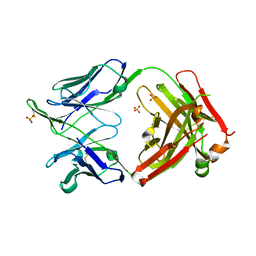 | | Crystal structure of broadly neutralizing antibody HEPC3 | | Descriptor: | SULFATE ION, antibody HEPC3 Heavy Chain, antibody HEPC3 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
6OKQ
 
 | | Crystal structure of the SF12 Fab | | Descriptor: | SF12 Fab Heavy Chain,SF12 Fab Heavy Chain, SF12 Fab Light Chain,SF12 Fab Light Chain | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2019-04-14 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope.
Immunity, 50, 2019
|
|
7TQG
 
 | | Crystal Structure of IOMA Fab inferred germline | | Descriptor: | IOMA iGL Fab Heavy Chain, IOMA iGL Fab Light Chain | | Authors: | Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | CD4 binding site immunogens elicit heterologous anti-HIV-1 neutralizing antibodies in transgenic and wild-type animals.
Sci Immunol, 8, 2023
|
|
7UZB
 
 | |
7UZC
 
 | |
7UZD
 
 | |
7UCF
 
 | |
7UCE
 
 | |
7UCG
 
 | |
7UZ9
 
 | |
7UZ8
 
 | |
