3V32
 
 | | Crystal structure of MCPIP1 N-terminal conserved domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Xu, J, Peng, W, Sun, Y, Wang, X, Xu, Y, Li, X, Gao, G, Rao, Z. | | Deposit date: | 2011-12-12 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase
Nucleic Acids Res., 40, 2012
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
6UM2
 
 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
3P6Y
 
 | | CF Im25-CF Im68-UGUAA complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
5TRD
 
 | | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*CP*TP*AP*AP*TP*TP*CP*AP*CP*GP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*CP*GP*TP*GP*AP*AP*TP*TP*AP*GP*TP*AP*A)-3'), ... | | Authors: | Vetting, M.W, Rodionova, I.A, Li, X, Osterman, A.L, Rodionov, D.A, Almo, S.C. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator
To be published
|
|
4MZD
 
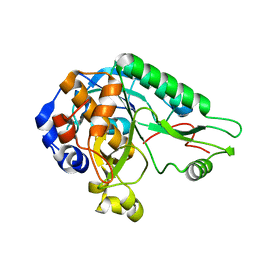 | | High resolution crystal structure of the nisin leader peptidase NisP from Lactococcus lactis | | Descriptor: | Nisin leader peptide-processing serine protease NisP | | Authors: | Rao, Z.H, Xu, Y.Y, Li, X, Yang, W. | | Deposit date: | 2013-09-30 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the nisin leader peptidase NisP revealing a C-terminal autocleavage activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6UOX
 
 | | Structure of itraconazole-bound NPC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-bromo-4-{4-[4-({(2R,4S)-2-(2,4-dichlorophenyl)-2-[(1H-1,2,4-triazol-1-yl)methyl]-1,3-dioxolan-4-yl}methoxy)phenyl]piperazin-1-yl}phenyl)-2-[(2S)-butan-2-yl]-2,4-dihydro-3H-1,2,4-triazol-3-one, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis for itraconazole-mediated NPC1 inhibition.
Nat Commun, 11, 2020
|
|
4QWQ
 
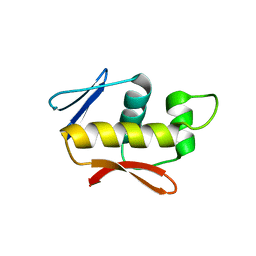 | | Crystal structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus | | Descriptor: | Response regulator SaeR | | Authors: | Fan, X, Zhu, Y, Zhang, X, Teng, M, Li, X. | | Deposit date: | 2014-07-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure of the DNA-binding domain of the response regulator SaeR from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4A1G
 
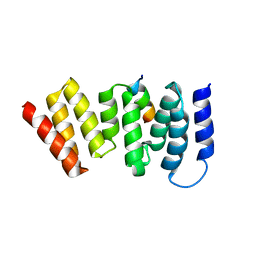 | | The crystal structure of the human Bub1 TPR domain in complex with the KI motif of Knl1 | | Descriptor: | MITOTIC CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, PROTEIN CASC5 | | Authors: | Krenn, V, Wehenkel, A, Li, X, Santaguida, S, Musacchio, A. | | Deposit date: | 2011-09-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis Reveals Features of the Spindle Checkpoint Kinase Bub1-Kinetochore Subunit Knl1 Interaction.
J.Cell Biol., 196, 2012
|
|
1C9W
 
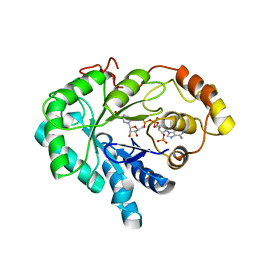 | | CHO REDUCTASE WITH NADP+ | | Descriptor: | CHO REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Li, X, Hyndman, D, Flynn, T.G, Jia, Z. | | Deposit date: | 1999-08-03 | | Release date: | 2000-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
Proteins, 38, 2000
|
|
3PUN
 
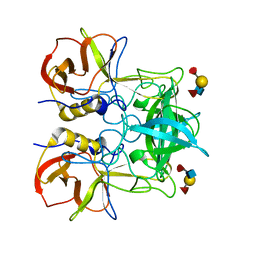 | | Crystal structure of P domain dimer of Norovirus VA207 with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3PVD
 
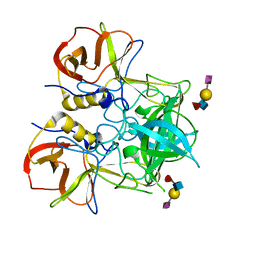 | | Crystal structure of P domain dimer of Norovirus VA207 complexed with 3'-sialyl-Lewis x tetrasaccharide | | Descriptor: | Capsid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3EZR
 
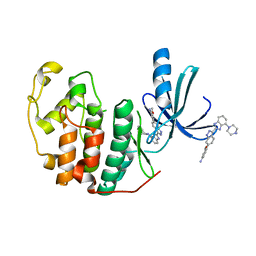 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3F5X
 
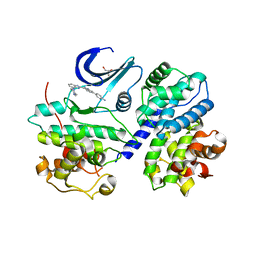 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EZV
 
 | | CDK-2 with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3QXL
 
 | | Crystal structure of the CDC25 Domain from Ral-specific Guanine-nucleotide Exchange Factor RalGPS1a | | Descriptor: | Ras-specific guanine nucleotide-releasing factor RalGPS1 | | Authors: | Peng, W, Xu, J, Guan, X, Sun, Y, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2011-03-02 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural study of the Cdc25 domain from Ral-specific guanine-nucleotide exchange factor RalGPS1a.
Protein Cell, 2, 2011
|
|
6AYG
 
 | | Human Apo-TRPML3 channel at pH 4.8 | | Descriptor: | Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6AYE
 
 | | Human apo-TRPML3 channel at pH 7.4 | | Descriptor: | Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2H0P
 
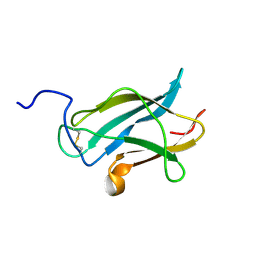 | | NMR Structure of the Dengue-4 virus Envelope Protein Domain III | | Descriptor: | Envelope glycoprotein | | Authors: | Volk, D.E, Lee, Y, Li, X, Thiviyanathan, V, Barrett, A.D.T, Gorenstein, D.G. | | Deposit date: | 2006-05-15 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the envelope protein domain III of dengue-4 virus.
Virology, 364, 2007
|
|
6AYF
 
 | | TRPML3/ML-SA1 complex at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3PUM
 
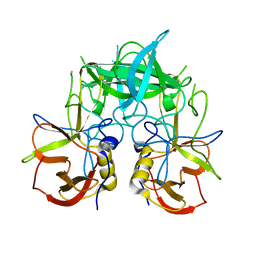 | | Crystal structure of P domain dimer of Norovirus VA207 | | Descriptor: | Capsid | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
2OBS
 
 | |
7Y1Q
 
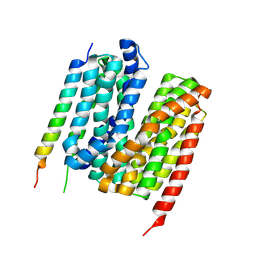 | | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | | Descriptor: | Isoform 2 of Basigin, Monocarboxylate transporter 1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.03 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2
To Be Published
|
|
7Y1B
 
 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
