7EU8
 
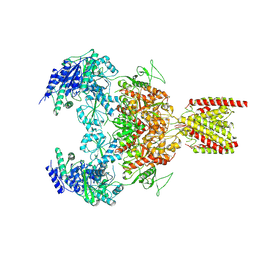 | | Structure of the human GluN1-GluN2B NMDA receptor in complex with S-ketamine,glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, T, Zhang, Y, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
7EU7
 
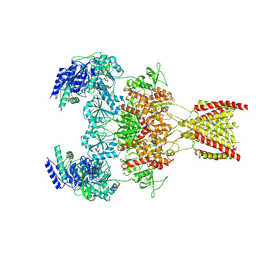 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
3DM7
 
 | |
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
3UO7
 
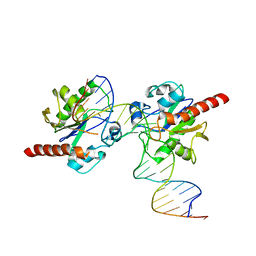 | | Crystal structure of Human Thymine DNA Glycosylase Bound to Substrate 5-carboxylcytosine | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*AP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(1CC)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | Authors: | Zhang, L, He, C. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.
Nat.Chem.Biol., 8, 2012
|
|
3UOB
 
 | |
6JV3
 
 | |
6JV5
 
 | | Crystal structure of 5-methylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(5CM)P*GP*CP*TP*GP*G)-3') | | Authors: | Zhang, L, Wang, Y.X. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
4QKN
 
 | | Crystal structure of FTO bound to a selective inhibitor | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2014-06-07 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5.
Nucleic Acids Res., 43, 2015
|
|
6LG9
 
 | |
6LG8
 
 | |
2Q15
 
 | | Structure of BACE complexed to compound 3a | | Descriptor: | (4S)-4-(2-AMINO-6-PHENOXYQUINAZOLIN-3(4H)-YL)-N,4-DICYCLOHEXYL-N-METHYLBUTANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
2Q11
 
 | | Structure of BACE complexed to compound 1 | | Descriptor: | 3-(2-AMINO-6-BENZOYLQUINAZOLIN-3(4H)-YL)-N-CYCLOHEXYL-N-METHYLPROPANAMIDE, Beta-secretase 1 | | Authors: | Sharff, A.J. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-3,4-dihydroquinazolines as inhibitors of BACE-1 (beta-Site APP cleaving enzyme): Use of structure based design to convert a micromolar hit into a nanomolar lead.
J.Med.Chem., 50, 2007
|
|
5TDX
 
 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
5LMK
 
 | | Structure of phopsho-CDK2-cyclin A in complex with an ATP-competitive inhibitor | | Descriptor: | 4-[4-[3-bromanyl-7-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-5-yl]phenyl]benzamide, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Explicit treatment of active-site waters enhances quantum mechanical/implicit solvent scoring: Inhibition of CDK2 by new pyrazolo[1,5-a]pyrimidines.
Eur J Med Chem, 126, 2016
|
|
1CXV
 
 | | STRUCTURE OF RECOMBINANT MOUSE COLLAGENASE-3 (MMP-13) | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, PROTEIN (COLLAGENASE-3), ... | | Authors: | Botos, I, Meyer, E, Swanson, S.M, Lemaitre, V, Eeckhout, Y, Meyer, E.F. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant mouse collagenase-3 (MMP-13).
J.Mol.Biol., 292, 1999
|
|
8OSB
 
 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
3SHI
 
 | | Crystal structure of human MMP1 catalytic domain at 2.2 A resolution | | Descriptor: | CALCIUM ION, Interstitial collagenase, ZINC ION | | Authors: | Bertini, I, Calderone, V, Cerofolini, L, Fragai, M, Geraldes, C.F.G.C, Hermann, P, Luchinat, C, Parigi, G, Teixeira, J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of MMP-1 studied through tagged lanthanides.
Febs Lett., 586, 2012
|
|
