5ADU
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R, Brooke, E.J, Wehlin, S.A, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4F
 
 | | The mechanism of Hydrogen Activation by NiFe-hydrogenases. | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4M
 
 | | Mechanism of Hydrogen activation by NiFe-hydrogenases | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sergent, F, Carr, S.B, Philips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
4UE3
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
5SXY
 
 | |
3CB4
 
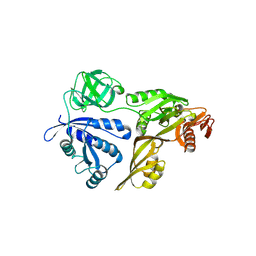 | | The Crystal Structure of LepA | | Descriptor: | GTP-binding protein lepA | | Authors: | Evans, R.N, Blaha, G, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-02-21 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of LepA, the ribosomal back translocase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
8EUO
 
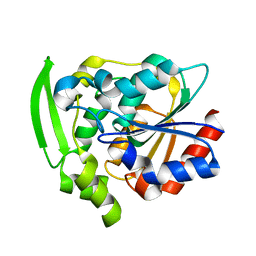 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
5LMM
 
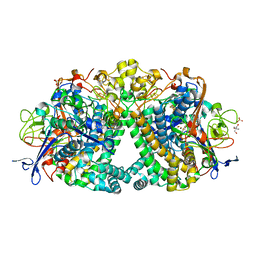 | | Structure of E coli Hydrogenase Hyd-1 mutant E28Q | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetic consequences of re-engineering the outer shell "canopy" above the active site of a [NiFe]-hydrogenase.
To Be Published
|
|
5LRY
 
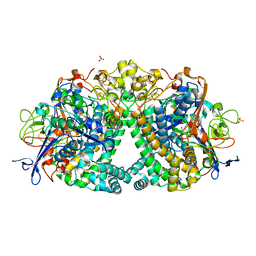 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
1Y0S
 
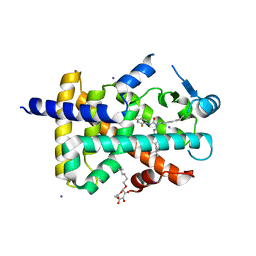 | | Crystal structure of PPAR delta complexed with GW2331 | | Descriptor: | (2S)-2-(4-[2-(3-[2,4-DIFLUOROPHENYL]-1-HEPTYLUREIDO)ETHYL]PHENOXY)-2-METHYLBUTYRIC ACID, IODIDE ION, Peroxisome proliferator activated receptor delta, ... | | Authors: | Takada, I, Yu, R.T, Xu, H.E, Xu, R.X, Lambert, M.H, Montana, V.G, Kliewer, S.A, Evans, R.M, Umesono, K. | | Deposit date: | 2004-11-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Alteration of a Single Amino Acid in Peroxisome Proliferator-Activated Receptor-alpha (PPARalpha) Generates a PPAR delta Phenotype
MOL.ENDOCRINOL., 14, 2000
|
|
1LO1
 
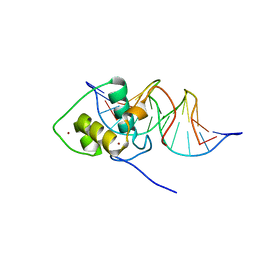 | | ESTROGEN RELATED RECEPTOR 2 DNA BINDING DOMAIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*CP*GP*TP*GP*AP*CP*CP*TP*TP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*AP*AP*GP*GP*TP*CP*AP*CP*G)-3', Steroid hormone receptor ERR2, ... | | Authors: | Gearhart, M.D, Holmbeck, S.M.A, Evans, R.M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-05-05 | | Release date: | 2003-04-22 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Monomeric Complex of Human Orphan Estrogen Related Receptor-2 with DNA: A Pseudo-dimer Interface Mediates Extended Half-site Recognition
J.Mol.Biol., 327, 2003
|
|
8SNI
 
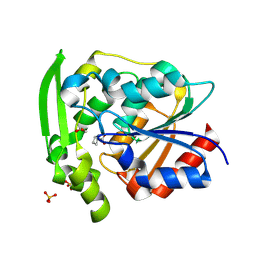 | | Hydroxynitrile Lyase from Hevea brasiliensis with Forty Mutations | | Descriptor: | (S)-hydroxynitrile lyase, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Walsh, M.E, Greenberg, L.R, Kazlauskas, R.J, Pierce, C.T, Aihara, H, Evans, R.L, Shi, K. | | Deposit date: | 2023-04-27 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | To be published
To Be Published
|
|
1OSH
 
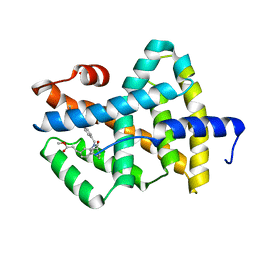 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
1KBH
 
 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
5TDX
 
 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
6SYO
 
 | | Hydrogenase-2 variant R479K - As Isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6SZK
 
 | | Hydrogenase-2 variant R479K - hydrogen reduced form treated with CO | | Descriptor: | CARBON MONOXIDE, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6SYX
 
 | | Hydrogenase-2 variant R479K - reduced sample exposed to pure oxygen | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6SZD
 
 | | Hydrogenase-2 variant R479K - hydrogen reduced form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
6EHS
 
 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EN9
 
 | | E. coli Hydrogenase-2 (hydrogen reduced form) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EHQ
 
 | | E. coli Hydrogenase-2 (as isolated form). | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6FPI
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
