2LCI
 
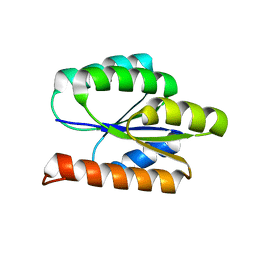 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN, P-LOOP NTPASE FOLD, Northeast Structural Genomics Consortium Target OR36 (CASD Target) | | Descriptor: | Protein OR36 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Janjua, H, Ciccosanti, C, Lee, H, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR36
To be Published
|
|
4RZ7
 
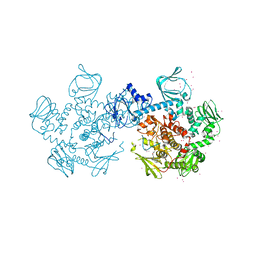 | | Crystal Structure of PVX_084705 with bound PCI32765 | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, UNKNOWN ATOM OR ION, cGMP-dependent protein kinase, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal Structure of PVX_084705 with bound PCI32765
To be Published
|
|
4NYX
 
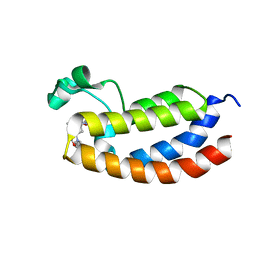 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand | | Descriptor: | (3R)-N-[3-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)propyl]-3-methyl-2-oxo-1,2,3,4-tetrahydroquinoxaline-5-carboxamide, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Rooney, T.P.C, Fedorov, O, Martin, S, Monteiro, O.P, Conway, S.J, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand
TO BE PUBLISHED
|
|
4NYV
 
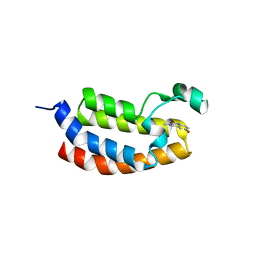 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a quinazolin-one ligand | | Descriptor: | 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, Martin, S, Monteiro, O.P, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a quinazolin-one ligand
TO BE PUBLISHED
|
|
1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | Descriptor: | MM1357 | | Authors: | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
3ITK
 
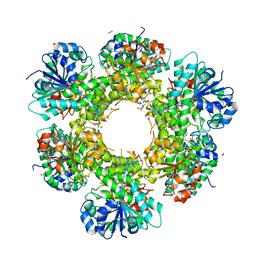 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
3EMW
 
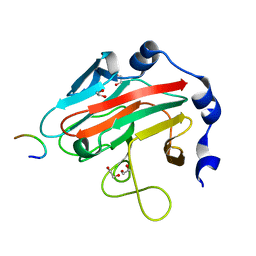 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) in complex with a 20-residue VASA peptide | | Descriptor: | 1,2-ETHANEDIOL, Peptide (VASA), SPRY domain-containing SOCS box protein 2 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A.K, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
3ERE
 
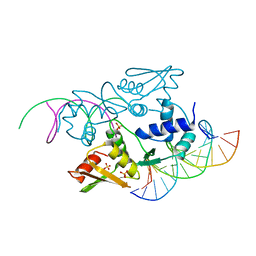 | | Crystal structure of the arginine repressor protein from Mycobacterium tuberculosis in complex with the DNA operator | | Descriptor: | 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DAP*DAP*DCP*DGP*DAP*DTP*DGP*DCP*DAP*DA)-3', 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DCP*DGP*DTP*DTP*DAP*DTP*DGP*DCP*DAP*DA)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis.
J.Mol.Biol., 384, 2008
|
|
2LEK
 
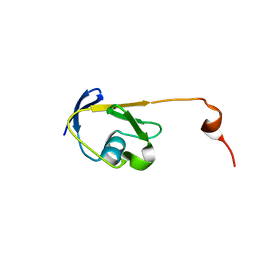 | | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris refined with NH RDCs. Northeast Structural Genomics Consortium target RpR325 | | Descriptor: | Putative thiamin biosynthesis ThiS | | Authors: | Ramelot, T.A, Cort, J.R, Lee, H, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium target RpR325
To be Published
|
|
2LSE
 
 | | Solution NMR Structure of De Novo Designed Four Helix Bundle Protein, Northeast Structural Genomics Consortium (NESG) Target OR188 | | Descriptor: | Four Helix Bundle Protein | | Authors: | Sathyamoorthy, B, Pulavarti, S, Murphy, G, Mills, J.L, Eletski, A, Der, B.S, Machius, M.C, Kuhlman, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Denovo Design Four Helix Bundle Protein, Northeast Structural Genomics Consortium Target OR188
To be Published
|
|
2JZ6
 
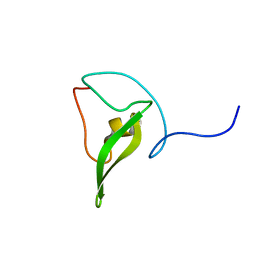 | | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97 | | Descriptor: | 50S ribosomal protein L28 | | Authors: | Wu, Y, Singarapu, K.K, Guido, V, Yee, A, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97.
To be Published
|
|
4MR4
 
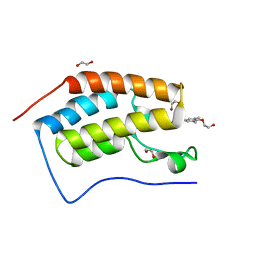 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2JXX
 
 | | NMR solution structure of Ubiquitin-like domain of NFATC2IP. Northeast Structural Genomics Consortium target HR5627 | | Descriptor: | NFATC2-interacting protein | | Authors: | Doherty, R.S, Dhe-Paganon, S, Fares, C, Lemak, A, Butler, C, Srisailam, S, Karra, M, Yee, A, Edwards, A.M, Weigelt, J, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Ubiquitin-like domain of NFATC2IP.
TO BE PUBLISHED
|
|
2L06
 
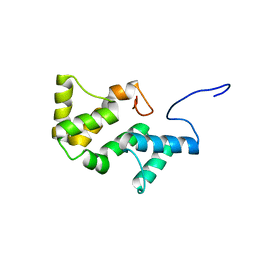 | | Solution NMR structure of the PBS linker polypeptide domain (fragment 254-400) of phycobilisome linker protein ApcE from Synechocystis sp. PCC 6803. Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Hamilton, K, Ciccosanti, C, Lee, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the PBS linker polypeptide domain of phycobilisome
linker protein apcE from Synechocystis sp. Northeast Structural Genomics Consortium
Target SgR209C
To be Published
|
|
1XN6
 
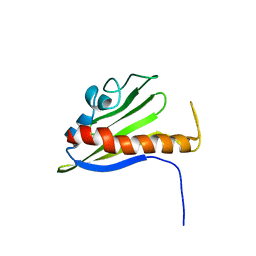 | | Solution Structure of Northeast Structural Genomics Target Protein BcR68 encoded in gene Q816V6 of B. cereus | | Descriptor: | hypothetical protein BC4709 | | Authors: | Liu, G, Acton, T, Parish, D, Ma, L, Xu, D, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3LF0
 
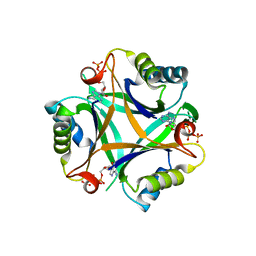 | | Crystal structure of the ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Shetty, N.D, Palaninathan, S.K, Reddy, M.C.M, Owen, J.L, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-15 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the apo and ATP bound Mycobacterium tuberculosis nitrogen regulatory PII protein.
Protein Sci., 19, 2010
|
|
2LLL
 
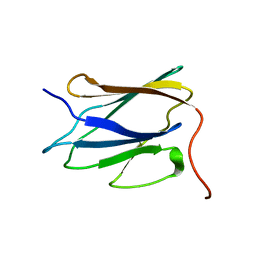 | | Solution NMR structure of C-terminal globular domain of human Lamin-B2, Northeast Structural Genomics Consortium target HR8546A | | Descriptor: | Lamin-B2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Xu, C, Min, J, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2011-11-11 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of c-terminal globular domain of human Lamin-B2
To be Published
|
|
3LAP
 
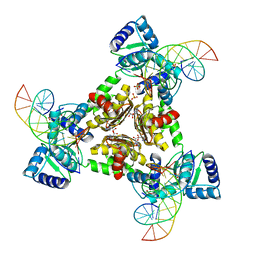 | | The Structure of the Intermediate Complex of the Arginine Repressor from Mycobacterium tuberculosis Bound to its DNA Operator and L-canavanine. | | Descriptor: | 5'-D(*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*A)-3', 5'-D(*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*A)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-01-06 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression.
J.Mol.Biol., 399, 2010
|
|
4NR6
 
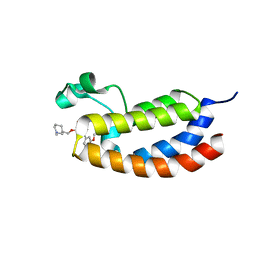 | | Crystal structure of the bromodomain of human CREBBP in complex with an oxazepin ligand | | Descriptor: | 1-[7-(3,4-dimethoxyphenyl)-9-{[(3R)-1-methylpiperidin-3-yl]methoxy}-2,3-dihydro-1,4-benzoxazepin-4(5H)-yl]propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Chaikuad, A, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an oxazepin ligand
To be Published
|
|
4NRB
 
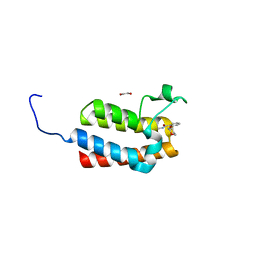 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-1 N01197 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Authors: | Muniz, J.R.C, Felletar, I, Chaikuad, A, Filippakopoulos, P, Ferguson, F.M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
2JOZ
 
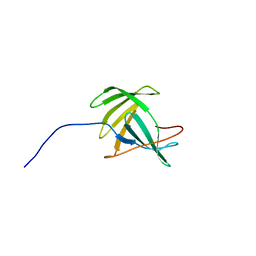 | | Solution NMR structure of protein yxeF, Northeast Structural Genomics Consortium target Sr500a | | Descriptor: | Hypothetical protein yxeF | | Authors: | Wu, Y, Liu, G, Zhang, Q, Bhatnagar, S, Chen, C, Nwosu, C, Xiao, R, Cunningham, K, Locke, J, Ma, L, Swapna, G, Baran, M, Acton, T, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein yxeF , Northeast Structural Genomics Consortium target Sr500a
To be Published
|
|
4NQN
 
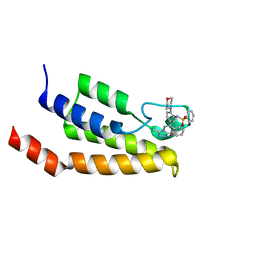 | | Crystal Structure of the bromodomain of human BRD9 in complex with a triazolo-phthalazine ligand | | Descriptor: | 2-chloro-N-[5-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-2-(morpholin-4-yl)phenyl]benzenesulfonamide, Bromodomain-containing protein 9 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal Structure of the bromodomain of human BRD9 in complex with a triazolo-phthalazine ligand
To be Published
|
|
2L82
 
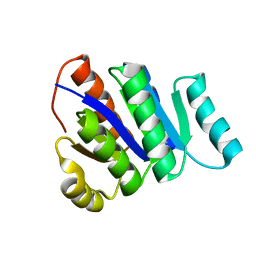 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR32 | | Descriptor: | DESIGNED PROTEIN OR32 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Janjua, H, Tong, S, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR32
To be Published
|
|
2LNI
 
 | | Solution NMR Structure of Stress-induced-phosphoprotein 1 STI1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4403E | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Tang, Y, Liu, G, Hamilton, K, Ciccosanti, C, Shastry, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4403E
To be Published
|
|
3U13
 
 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
