5VHS
 
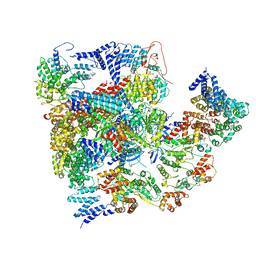 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHF
 
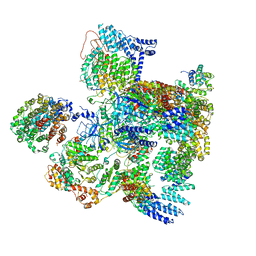 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHH
 
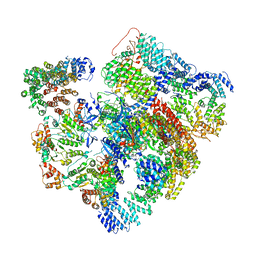 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHI
 
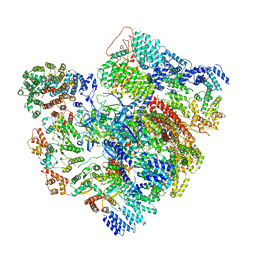 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
7SIM
 
 | | Structure of positive allosteric modulator-free active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2XCL
 
 | |
2XD4
 
 | |
1H69
 
 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
5A8H
 
 | | cryo-ET subtomogram averaging of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195 VARIANT G52K5, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-15 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env.
Cell, 162, 2015
|
|
5A7X
 
 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
4NFT
 
 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | Authors: | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | Deposit date: | 2013-11-01 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
4J1L
 
 | | Mutant Endotoxin TeNT | | Descriptor: | Tetanus toxin, ZINC ION | | Authors: | Guo, J, Pan, X, Zhao, Y, Chen, S. | | Deposit date: | 2013-02-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering Clostridia Neurotoxins with elevated catalytic activity
Toxicon, 74, 2013
|
|
5FNA
 
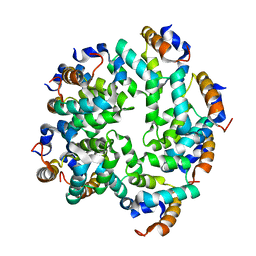 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
3TCM
 
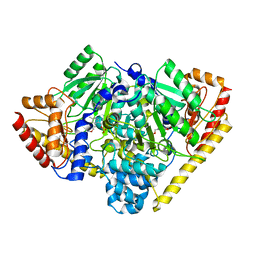 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
8KDB
 
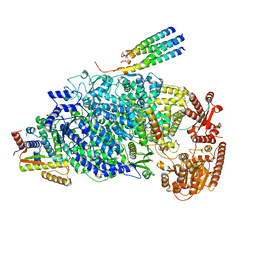 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8KDC
 
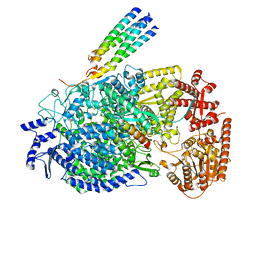 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
4GHQ
 
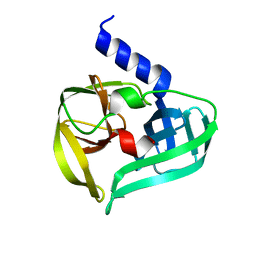 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5C20
 
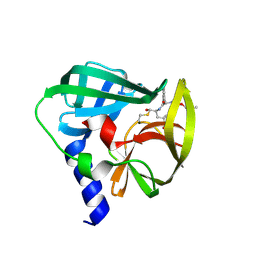 | | Crystal structure of EV71 3C Proteinase in complex with Compound 2 | | Descriptor: | 2-methylpropyl N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1U
 
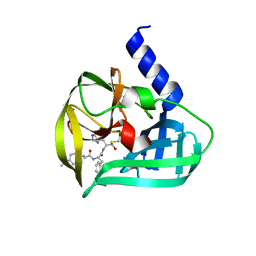 | | Crystal structure of EV71 3C Proteinase in complex with Compound Xb | | Descriptor: | (2S)-2-[[(E)-3-[4-(dimethylamino)phenyl]prop-2-enoyl]amino]-N-[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-phenyl-propanamide, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
5C1X
 
 | | Crystal structure of EV71 3C Proteinase in complex with Compound VIII | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C proteinase | | Authors: | Zhang, L, Huang, G, Cai, Q, Zhao, C, Ren, H, Li, P, Li, N, Chen, S, Li, J, Lin, T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Optimize the interactions at S4 with efficient inhibitors targeting 3C proteinase from enterovirus 71
J.Mol.Recognit., 29, 2016
|
|
4GP3
 
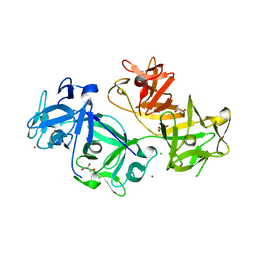 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
