8HK2
 
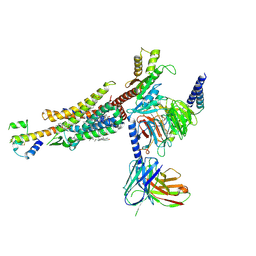 | | C3aR-Gi-C3a protein complex | | 分子名称: | C3a anaphylatoxin, C3a anaphylatoxin chemotactic receptor, CHOLESTEROL, ... | | 著者 | Wang, Y, Liu, W, Xu, Y, Zhuang, Y, Xu, H.E. | | 登録日 | 2022-11-24 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins.
Nat.Chem.Biol., 19, 2023
|
|
5YAA
 
 | |
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | 分子名称: | Methyltransferase | | 著者 | Chen, H, Lin, S, Lu, G.W. | | 登録日 | 2022-09-21 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
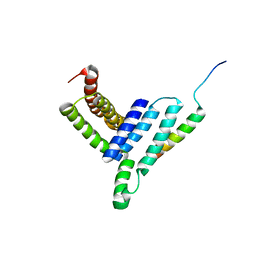
5H7P
 
 | |
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | 著者 | Yang, J, Lin, S, Lu, G.W. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8HQ6
 
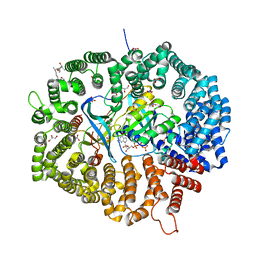 | | KL2 in complex with CRM1-Ran-RanBP1 | | 分子名称: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | 著者 | Sun, Q, Jian, L. | | 登録日 | 2022-12-13 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8H5T
 
 | |
8HQ3
 
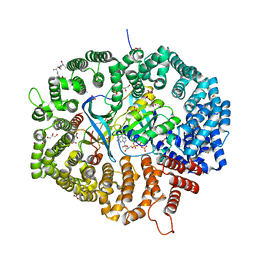 | | KL1 in complex with CRM1-Ran-RanBP1 | | 分子名称: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | 著者 | Sun, Q, Jian, L. | | 登録日 | 2022-12-13 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
2FDY
 
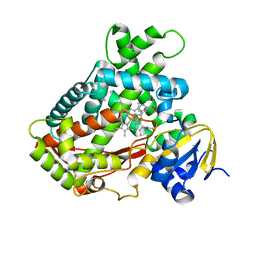 | | Microsomal P450 2A6 with the inhibitor Adrithiol bound | | 分子名称: | 4,4'-DIPYRIDYL DISULFIDE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
8W8K
 
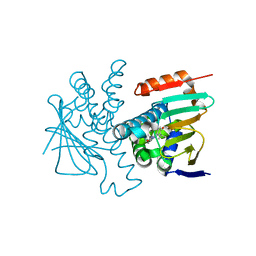 | | Crystal structures of HSP90 and the compound Ganetespid in the "closed" conformation | | 分子名称: | 5-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-(1-methyl-1H-indol-5-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | 著者 | Xu, C, Zhang, X.L, Bai, F. | | 登録日 | 2023-09-03 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
8W4V
 
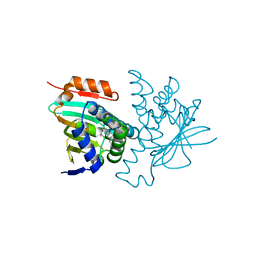 | | Crystal structure of human HSP90 in complex with compound 4 | | 分子名称: | 4-[2-[(dimethylamino)methyl]phenyl]sulfanylbenzene-1,3-diol, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | 著者 | Xu, C, Zhang, X.L, Bai, F. | | 登録日 | 2023-08-25 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
4LA2
 
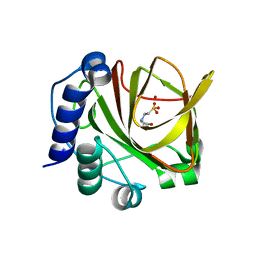 | |
5HZ6
 
 | |
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
8HVS
 
 | |
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | 分子名称: | Major capsid protein, UL17, UL25, ... | | 著者 | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-05-31 | | 公開日 | 2018-10-10 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
5HZ8
 
 | | FABP4_3 in complex with 6,8-Dichloro-4-phenyl-2-piperidin-1-yl-quinoline-3-carboxylic acid | | 分子名称: | 6,8-dichloro-4-phenyl-2-(piperidin-1-yl)quinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Rudolph, M.G. | | 登録日 | 2016-02-02 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Design and synthesis of selective, dual fatty acid binding protein 4 and 5 inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5HZ9
 
 | |
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | 著者 | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2018-03-24 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | 分子名称: | CidA_I gamma/2 protein, CidB_I b/2 protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | 分子名称: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | 分子名称: | ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
