4R05
 
 | | Crystal structure of the refolded DENV3 methyltransferase | | 分子名称: | Nonstructural protein NS5 | | 著者 | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | 登録日 | 2014-07-29 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
8J36
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF00835231 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zhou, X.L, Lin, C, Zou, X.F, Zhang, J, Li, J. | | 登録日 | 2023-04-16 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Crystal structure of SARS-Cov-2 main protease
M49I mutant in complex with PF00835231
To Be Published
|
|
6LXD
 
 | | Pri-miRNA bound DROSHA-DGCR8 complex | | 分子名称: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | 著者 | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | 登録日 | 2020-02-10 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
4E9B
 
 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with actinonin | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACTINONIN, COBALT (II) ION, ... | | 著者 | Cui, K, Zhu, L, Lu, W, Huang, J. | | 登録日 | 2012-03-20 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | 分子名称: | E protein | | 著者 | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | 登録日 | 2017-01-14 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6LXE
 
 | | DROSHA-DGCR8 complex | | 分子名称: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | 著者 | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | 登録日 | 2020-02-10 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
2LGX
 
 | | NMR structure for Kindle-2 N-terminus | | 分子名称: | Fermitin family homolog 2 | | 著者 | Perera, H.D, Ma, Y, Yang, J, Hirbawi, J, Plow, E.F, Qin, J. | | 登録日 | 2011-08-03 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Membrane Binding of the N-Terminal Ubiquitin-Like Domain of kindlin-2 Is Crucial for Its Regulation of Integrin Activation.
Structure, 19, 2011
|
|
3HIF
 
 | | The crystal structure of apo wild type CAP at 3.6 A resolution. | | 分子名称: | Catabolite gene activator | | 著者 | Steitz, T.A, Sharma, H, Wang, J, Kong, J, Yu, S. | | 登録日 | 2009-05-19 | | 公開日 | 2009-09-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8WGF
 
 | | The Crystal Structure of JNK3 from Biortus. | | 分子名称: | MAGNESIUM ION, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | 登録日 | 2023-09-21 | | 公開日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Crystal Structure of JNK3 from Biortus.
To Be Published
|
|
8WGQ
 
 | | The Crystal Structure of L-asparaginase from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, L-asparaginase | | 著者 | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | 登録日 | 2023-09-22 | | 公開日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Crystal Structure of L-asparaginase from Biortus.
To Be Published
|
|
3HRO
 
 | | Crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | 分子名称: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2), also called Polycystin-2 or polycystic kidney disease 2(PKD2) | | 著者 | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | 登録日 | 2009-06-09 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7K65
 
 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | 著者 | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | 分子名称: | Solute carrier family 22 member 6 | | 著者 | Dou, T, Jiang, J. | | 登録日 | 2023-04-07 | | 公開日 | 2023-10-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.05 Å) | | 主引用文献 | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SDZ
 
 | |
7DR8
 
 | | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121 | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-26 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.338149 Å) | | 主引用文献 | Crystal structure of MERS-CoV 3CL protease in spacegroup P212121
To Be Published
|
|
8SDY
 
 | |
3HRN
 
 | | crystal structure of a C-terminal coiled coil domain of Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2, polycystic kidney disease 2) | | 分子名称: | Transient receptor potential (TRP) channel subfamily P member 2 (TRPP2) | | 著者 | Yu, Y, Ulbrich, M.H, Li, M.-H, Buraei, Z, Chen, X.-Z, Ong, A.C.M, Tong, L, Isacoff, E.Y, Yang, J. | | 登録日 | 2009-06-09 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and molecular basis of the assembly of the TRPP2/PKD1 complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5G2X
 
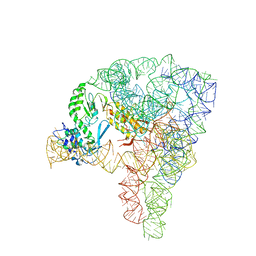 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | 分子名称: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | 著者 | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | 登録日 | 2016-04-16 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5G2Y
 
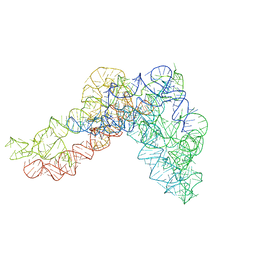 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | 分子名称: | GROUP II INTRON | | 著者 | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | 登録日 | 2016-04-16 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | 著者 | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | 登録日 | 2014-08-02 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
3IZJ
 
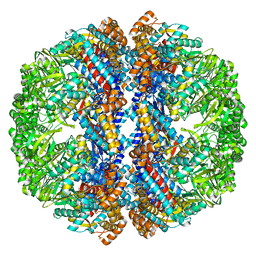 | | Mm-cpn rls with ATP and AlFx | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
5H0S
 
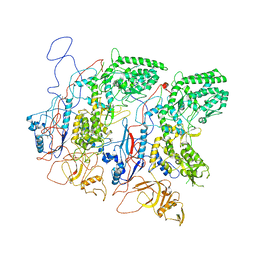 | | EM Structure of VP1A and VP1B | | 分子名称: | VP1 | | 著者 | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | 登録日 | 2016-10-06 | | 公開日 | 2017-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
4N0G
 
 | | Crystal Structure of PYL13-PP2CA complex | | 分子名称: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | 著者 | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | 登録日 | 2013-10-01 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
4FGB
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase I apo form | | 分子名称: | Calcium/calmodulin-dependent protein kinase type 1 | | 著者 | Zha, M, Zhong, C, Ou, Y, Wang, J, Han, L, Ding, J. | | 登録日 | 2012-06-04 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of human CaMKIalpha reveal insights into the regulation mechanism of CaMKI.
Plos One, 7, 2012
|
|
7DVM
 
 | |
