6L6R
 
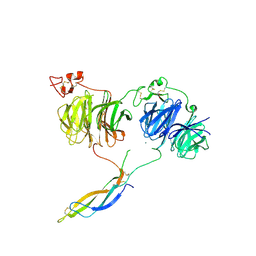 | | Crystal structure of LRP6 E1E2-SOST complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Choi, H.-J, Kim, J. | | 登録日 | 2019-10-29 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains.
Nat Commun, 11, 2020
|
|
3HWC
 
 | |
3K87
 
 | |
5TQM
 
 | | Cinnamoyl-CoA Reductase 1 from Sorghum bicolor in complex with NADP+ | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cinnamoyl-CoA Reductase, GLYCEROL, ... | | 著者 | Sattler, S.A, Kang, C.H. | | 登録日 | 2016-10-24 | | 公開日 | 2017-04-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Biochemical Characterization of Cinnamoyl-CoA Reductases.
Plant Physiol., 173, 2017
|
|
5KHT
 
 | | Crystal structure of the N-terminal fragment of tropomyosin isoform Tpm1.1 at 1.5 A resolution | | 分子名称: | DI(HYDROXYETHYL)ETHER, Tropomyosin alpha-1 chain,General control protein GCN4 | | 著者 | Kostyukova, A.S, Krieger, I, Yoon, Y.-H, Tolkatchev, D, Samatey, F.A. | | 登録日 | 2016-06-15 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.4964 Å) | | 主引用文献 | Structural destabilization of tropomyosin induced by the cardiomyopathy-linked mutation R21H.
Protein Sci., 27, 2018
|
|
7F0S
 
 | |
6L4Z
 
 | |
6L50
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | 分子名称: | 2-sulfanylidene-1,3-thiazolidin-4-one, NS3 protease, Serine protease subunit NS2B | | 著者 | Quek, J.P. | | 登録日 | 2019-10-21 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
4PPI
 
 | | Crystal structure of Bcl-xL hexamer | | 分子名称: | Bcl-2-like protein 1, GLYCEROL | | 著者 | Sreekanth, R, Yoon, H.S. | | 登録日 | 2014-02-27 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | 分子名称: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | 著者 | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | 登録日 | 2010-03-10 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | 分子名称: | Single-stranded DNA-binding protein | | 著者 | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | 登録日 | 2010-03-10 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5DQE
 
 | |
5DQ8
 
 | |
5GJ4
 
 | |
5H4I
 
 | |
5GPI
 
 | |
5H6V
 
 | |
5YOF
 
 | |
5YOD
 
 | |
7VW5
 
 | |
7VB4
 
 | |
7F8G
 
 | |
7F8H
 
 | |
2MHF
 
 | |
2LAV
 
 | |
