8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-25 | | 公開日 | 2023-06-28 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | 分子名称: | Spike protein S1, scFv | | 著者 | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | 登録日 | 2022-08-27 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
4LWH
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ5 | | 分子名称: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(propan-2-yl)phenyl]-4-[4-(morpholin-4-ylmethyl)phenyl]-1,2-oxazol-5-yl}cyclopropanecarboxamide | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
4LWE
 
 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ2 | | 分子名称: | Heat shock protein HSP 90-alpha, N-[3-(5-chloro-2,4-dihydroxyphenyl)-4-(4-methoxyphenyl)-1,2-oxazol-5-yl]acetamide | | 著者 | Li, J, Shi, F, Xiong, B, He, J. | | 登録日 | 2013-07-27 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of potent N-(isoxazol-5-yl)amides as HSP90 inhibitors.
Eur.J.Med.Chem., 87, 2014
|
|
8HBK
 
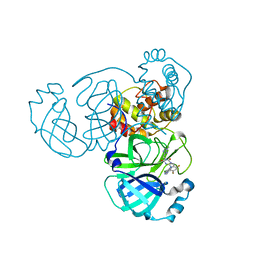 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M. | | 登録日 | 2022-10-29 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8WCW
 
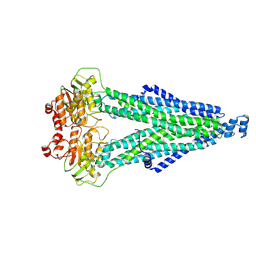 | |
8WCX
 
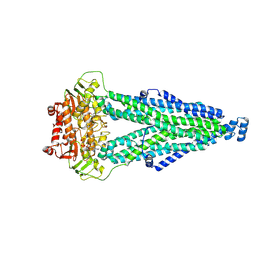 | |
1FJ1
 
 | |
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | 著者 | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | 登録日 | 2010-06-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGO
 
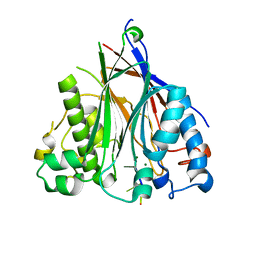 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | 分子名称: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | 著者 | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | 登録日 | 2010-06-12 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGQ
 
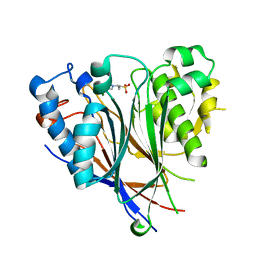 | | Crystal structure of the human CNOT6L nuclease domain | | 分子名称: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | 著者 | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | 登録日 | 2010-06-13 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
6LLG
 
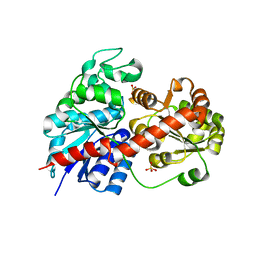 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | 分子名称: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | 著者 | Wang, X, Liu, M. | | 登録日 | 2019-12-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LLZ
 
 | |
7YW5
 
 | |
2OC7
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH571696 | | 分子名称: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, TERT-BUTYL {(1S)-2-[(1R,2S,5R)-2-({[(1S)-3-AMINO-1-(CYCLOBUTYLMETHYL)-2,3-DIOXOPROPYL]AMINO}CARBONYL)-7,7-DIMETHYL-6-OXA-3-AZABICYCLO[3.2.0]HEPT-3-YL]-1-CYCLOHEXYL-2-OXOETHYL}CARBAMATE, ... | | 著者 | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | 登録日 | 2006-12-20 | | 公開日 | 2007-07-31 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
4YWZ
 
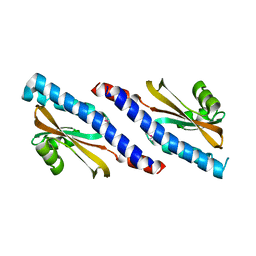 | |
6KVA
 
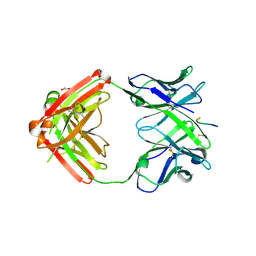 | | Structure of anti-hCXCR2 abN48-2 in complex with its CXCR2 epitope | | 分子名称: | 1,2-ETHANEDIOL, Peptide from C-X-C chemokine receptor type 2, heavy chain, ... | | 著者 | Xiang, J.C, Yan, L, Yang, B, Wilson, I.A. | | 登録日 | 2019-09-03 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Selection of a picomolar antibody that targets CXCR2-mediated neutrophil activation and alleviates EAE symptoms.
Nat Commun, 12, 2021
|
|
7FAP
 
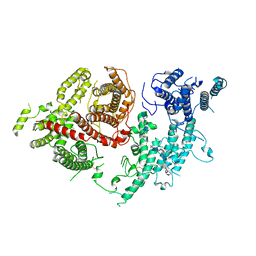 | | Structure of VAR2CSA-CSA 3D7 | | 分子名称: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | 著者 | Wang, L, Wang, Z. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
2P4L
 
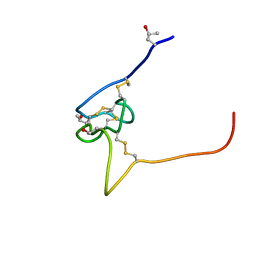 | | Structure and sodium channel activity of an excitatory I1-superfamily conotoxin | | 分子名称: | I-superfamily conotoxin r11a | | 著者 | Buczek, O, Wei, D.X, Babon, J.J, Yang, X.D, Fiedler, B, Yoshikami, D, Olivera, B.M, Bulaj, G, Norton, R.S. | | 登録日 | 2007-03-12 | | 公開日 | 2007-09-25 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Sodium Channel Activity of an Excitatory I(1)-Superfamily Conotoxin
Biochemistry, 46, 2007
|
|
6WJ5
 
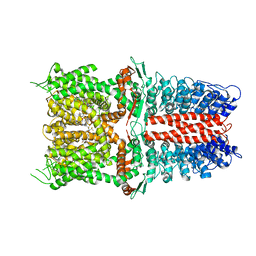 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | 分子名称: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | 著者 | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | 登録日 | 2020-04-11 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
6XOG
 
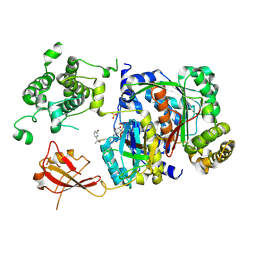 | | Structure of SUMO1-ML786519 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
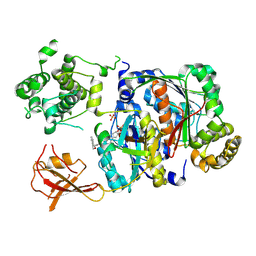 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6M2L
 
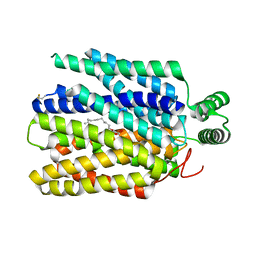 | | Crystal structure of Plasmodium falciparum hexose transporter PfHT1 bound with C3361 | | 分子名称: | (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Hexose transporter 1 | | 著者 | Jiang, X, Yuan, Y.Y, Zhang, S, Wang, N, Yan, C.Y, Yan, N. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural Basis for Blocking Sugar Uptake into the Malaria Parasite Plasmodium falciparum.
Cell, 183, 2020
|
|
