6OC9
 
 | |
7CYP
 
 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | 著者 | Wang, X, Zhu, L. | | 登録日 | 2020-09-04 | | 公開日 | 2021-06-09 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | 登録日 | 2020-10-16 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | 分子名称: | Major prion protein | | 著者 | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | 登録日 | 2021-01-18 | | 公開日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7F29
 
 | |
2AL5
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with fluoro-willardiine and aniracetam | | 分子名称: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2 | | 著者 | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
7VXG
 
 | |
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | 分子名称: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | 著者 | Lin, M, Paige, A, Tong, L. | | 登録日 | 2023-02-03 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
7C1D
 
 | |
5YAC
 
 | | Crystal structure of WT Trm5b from Pyrococcus abyssi | | 分子名称: | tRNA (guanine(37)-N1)-methyltransferase Trm5b | | 著者 | Xie, W, Wu, J. | | 登録日 | 2017-08-31 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | The crystal structure of the Pyrococcus abyssi mono-functional methyltransferase PaTrm5b
Biochem. Biophys. Res. Commun., 493, 2017
|
|
4ZLK
 
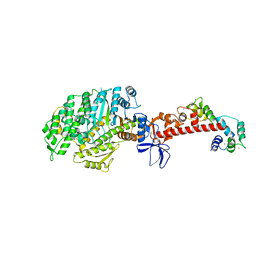 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | 分子名称: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | 著者 | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | 登録日 | 2015-05-01 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
8K20
 
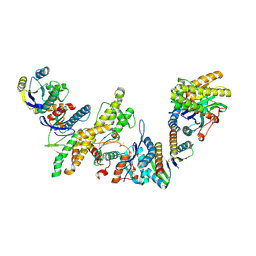 | | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | | 分子名称: | At4g34412, At5g53043, FE (III) ION, ... | | 著者 | Zheng, X.X, Zhu, L, Duan, L, Zhang, W.H. | | 登録日 | 2023-07-11 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Nucleic Acids Res., 52, 2024
|
|
7E3J
 
 | |
7F3B
 
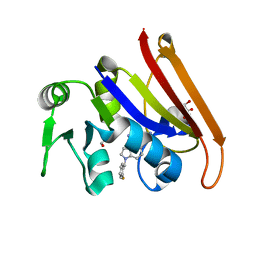 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | 分子名称: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | 著者 | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | 登録日 | 2021-06-16 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
7VLN
 
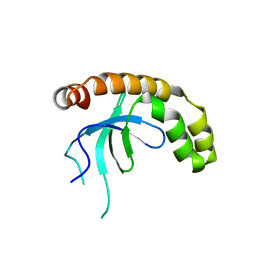 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | 分子名称: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | 著者 | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | 登録日 | 2021-10-05 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Li, L, Liao, H, Meng, Y, Li, W. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Li, W, Meng, Y, Liao, H. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liao, H, Meng, Y, Li, W. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
6KU8
 
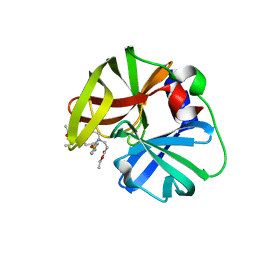 | | structure of HRV-C 3C protein with rupintrivir | | 分子名称: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, Genome polyprotein | | 著者 | Zhu, L, Yuan, S. | | 登録日 | 2019-08-31 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the HRV-C 3C-Rupintrivir Complex Provides New Insights for Inhibitor Design.
Virol Sin, 35, 2020
|
|
6KQX
 
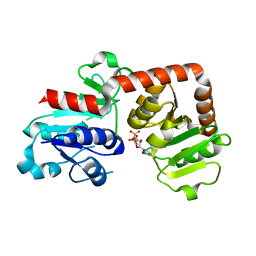 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | 分子名称: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | 著者 | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
2I3V
 
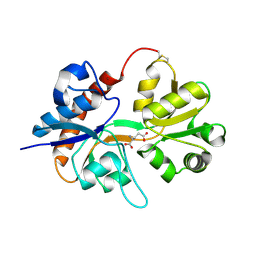 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | 著者 | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | 登録日 | 2006-08-21 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
2KN1
 
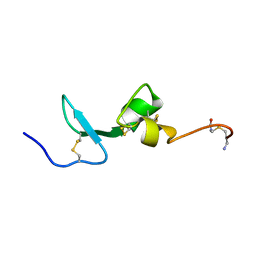 | | Solution NMR Structure of BCMA | | 分子名称: | Tumor necrosis factor receptor superfamily member 17 | | 著者 | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | 登録日 | 2009-08-11 | | 公開日 | 2011-02-23 | | 最終更新日 | 2013-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KN0
 
 | | Solution NMR Structure of xenopus Fn14 | | 分子名称: | Fn14 | | 著者 | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | 登録日 | 2009-08-11 | | 公開日 | 2011-06-29 | | 最終更新日 | 2013-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KMZ
 
 | | NMR Structure of hFn14 | | 分子名称: | Tumor necrosis factor receptor superfamily member 12A | | 著者 | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | 登録日 | 2009-08-10 | | 公開日 | 2011-06-29 | | 最終更新日 | 2013-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2I3W
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | 分子名称: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | 著者 | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | 登録日 | 2006-08-21 | | 公開日 | 2006-10-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
