8J5U
 
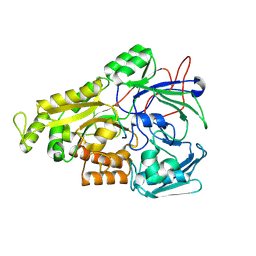 | | Crystal structure of Mycobacterium tuberculosis OppA complexed with an endogenous oligopeptide | | 分子名称: | Endogenous oligopeptide, Uncharacterized protein Rv1280c | | 著者 | Yang, X, Hu, T, Zhang, B, Rao, Z. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5S
 
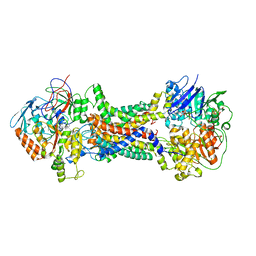 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-catalytic intermediate state | | 分子名称: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Yang, X, Hu, T, Zhang, B, Rao, Z. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5R
 
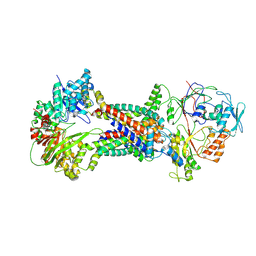 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the resting state | | 分子名称: | IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, Putative peptide transport permease protein Rv1283c, ... | | 著者 | Yang, X, Hu, T, Zhang, B, Rao, Z. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5T
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the catalytic intermediate state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Yang, X, Hu, T, Zhang, B, Rao, Z. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
8J5Q
 
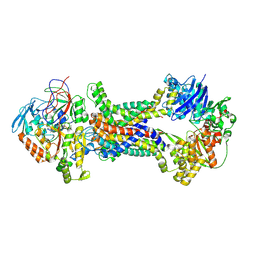 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-translocation state | | 分子名称: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, ... | | 著者 | Yang, X, Hu, T, Zhang, B, Rao, Z. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 2024
|
|
3CVA
 
 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | 分子名称: | Apoptosis regulator Bcl-X | | 著者 | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | 登録日 | 2008-04-18 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
3DOO
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis complexed with shikimate | | 分子名称: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Shikimate dehydrogenase | | 著者 | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | 登録日 | 2008-07-05 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
3DON
 
 | | Crystal structure of shikimate dehydrogenase from Staphylococcus epidermidis | | 分子名称: | GLYCEROL, Shikimate dehydrogenase | | 著者 | Han, C, Hu, T, Wu, D, Zhou, J, Shen, X, Qu, D, Jiang, H. | | 登録日 | 2008-07-05 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | X-ray crystallographic and enzymatic analyses of shikimate dehydrogenase from Staphylococcus epidermidis
Febs J., 276, 2009
|
|
7C2M
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | 著者 | Zhang, B, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-05-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7CAF
 
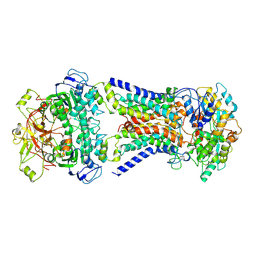 | | Mycobacterium smegmatis LpqY-SugABC complex in the pre-translocation state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7C2N
 
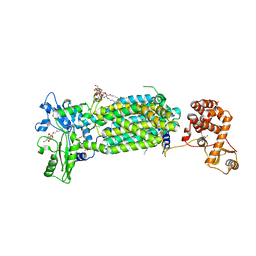 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | 著者 | Zhang, B, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-05-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7CAG
 
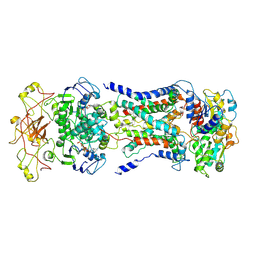 | | Mycobacterium smegmatis LpqY-SugABC complex in the catalytic intermediate state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7CAD
 
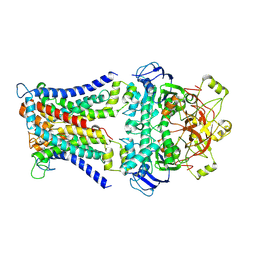 | | Mycobacterium smegmatis SugABC complex | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7CAE
 
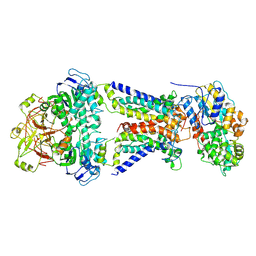 | | Mycobacterium smegmatis LpqY-SugABC complex in the resting state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
5C94
 
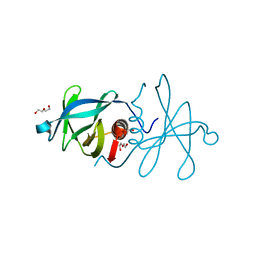 | | Infectious bronchitis virus nsp9 | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Non-structural protein 9 | | 著者 | Chen, C, Dou, Y, Yang, H, Su, D. | | 登録日 | 2015-06-26 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.438 Å) | | 主引用文献 | Structural basis for dimerization and RNA binding of avian infectious bronchitis virus nsp9.
Protein Sci., 26, 2017
|
|
4RE1
 
 | |
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6IZH
 
 | |
6WJI
 
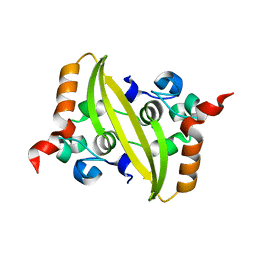 | | 2.05 Angstrom Resolution Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2 | | 分子名称: | CHLORIDE ION, Nucleoprotein | | 著者 | Minasov, G, Shuvalova, L, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-13 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Serodominant SARS-CoV-2 Nucleocapsid Peptides Map to Unstructured Protein Regions.
Microbiol Spectr, 2023
|
|
8J5Z
 
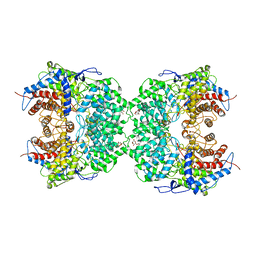 | | The cryo-EM structure of the TwOSC1 tetramer | | 分子名称: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | 著者 | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | 登録日 | 2023-04-24 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (4.75 Å) | | 主引用文献 | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3NSQ
 
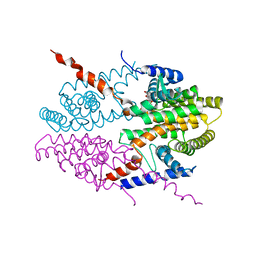 | | Crystal structure of tetrameric RXRalpha-LBD complexed with antagonist danthron | | 分子名称: | 1,8-dihydroxyanthracene-9,10-dione, Retinoid X receptor, alpha | | 著者 | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
3NSP
 
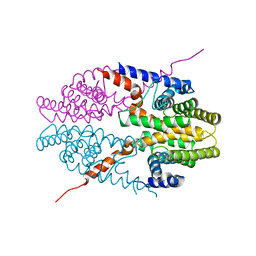 | | Crystal structure of tetrameric RXRalpha-LBD | | 分子名称: | Retinoid X receptor, alpha | | 著者 | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | 登録日 | 2010-07-02 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
6UYC
 
 | | Crystal structure of TEAD2 bound to Compound 2 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | 著者 | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | 登録日 | 2019-11-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.658 Å) | | 主引用文献 | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6UYB
 
 | | Crystal structure of TEAD2 bound to Compound 1 | | 分子名称: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | 著者 | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | 登録日 | 2019-11-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.543 Å) | | 主引用文献 | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-03-16 | | 公開日 | 2020-04-01 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
