3MOR
 
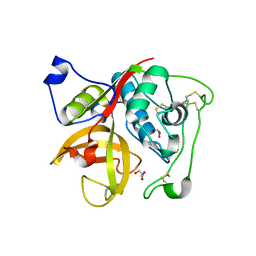 | | Crystal structure of Cathepsin B from Trypanosoma Brucei | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin B-like cysteine protease, ... | | 著者 | Cupelli, K, Stehle, T. | | 登録日 | 2010-04-23 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | In vivo protein crystallization opens new routes in structural biology.
Nat.Methods, 9, 2012
|
|
5J4E
 
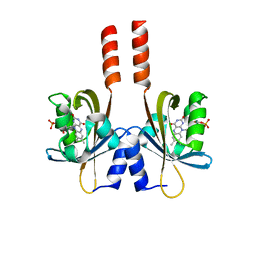 | |
5J3W
 
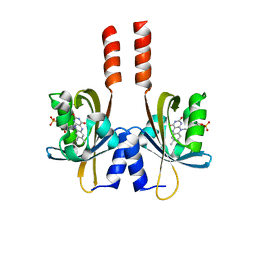 | |
1GKG
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | 分子名称: | COMPLEMENT RECEPTOR TYPE 1 | | 著者 | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | 登録日 | 2001-08-14 | | 公開日 | 2002-04-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
6ZTD
 
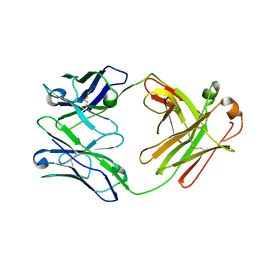 | |
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | 登録日 | 2012-05-29 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (4.9228 Å) | | 主引用文献 | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
5FV5
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | 分子名称: | ACETATE ION, Flocculation protein FLO11 | | 著者 | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
5FV6
 
 | | KpFlo11 presents a novel member of the Flo11 family with a unique recognition pattern for homophilic interactions | | 分子名称: | ACETATE ION, Flocculation protein FLO11, GLYCEROL, ... | | 著者 | Kraushaar, T, Brueckner, S, Mikolaiski, M, Schreiner, F, Veelders, M, Moesch, H.U, Essen, L.O. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-22 | | 最終更新日 | 2021-06-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family.
Elife, 9, 2020
|
|
3TG6
 
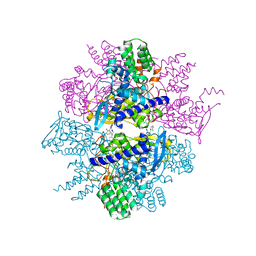 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | 分子名称: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | 著者 | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | 登録日 | 2011-08-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
3RO5
 
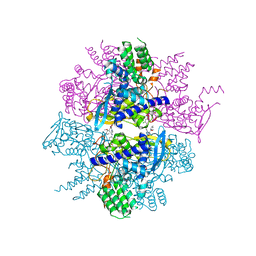 | | Crystal structure of influenza A virus nucleoprotein with ligand | | 分子名称: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | 著者 | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | 登録日 | 2011-04-25 | | 公開日 | 2011-09-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1A2P
 
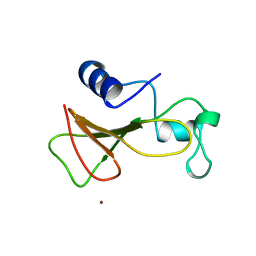 | | BARNASE WILDTYPE STRUCTURE AT 1.5 ANGSTROMS RESOLUTION | | 分子名称: | BARNASE, ZINC ION | | 著者 | Martin, C, Richard, V, Salem, M, Hartley, R.W, Mauguen, Y. | | 登録日 | 1998-01-07 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Refinement and structural analysis of barnase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
7UCW
 
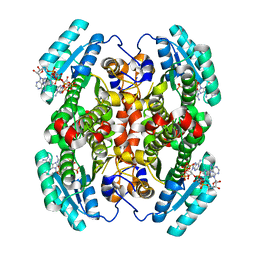 | | Structure of mouse Decr1 in complex with 2'-5' oligoadenylate | | 分子名称: | Decr1 protein, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | 著者 | Govande, A.A, Kranzusch, P.J. | | 登録日 | 2022-03-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | RNase L-activating 2'-5' oligoadenylates bind ABCF1, ABCF3 and Decr-1.
J.Gen.Virol., 104, 2023
|
|
5IFH
 
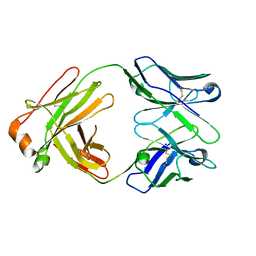 | | Crystal structure of the BCR Fab fragment from subset #2 case P11475 | | 分子名称: | Heavy chain of the Fab fragment from BCR derived from the P11475 CLL clone, Light chain of the Fab fragment from BCR derived from the P11475 CLL clone | | 著者 | Minici, C, Degano, M. | | 登録日 | 2016-02-26 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.293 Å) | | 主引用文献 | Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia.
Nat Commun, 8, 2017
|
|
2MCY
 
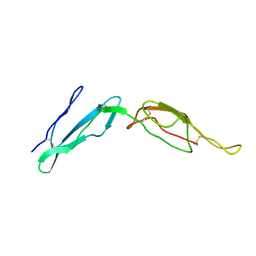 | | CR1 Sushi domains 2 and 3 | | 分子名称: | Complement receptor type 1 | | 著者 | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | 登録日 | 2013-08-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2014-01-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
2MCZ
 
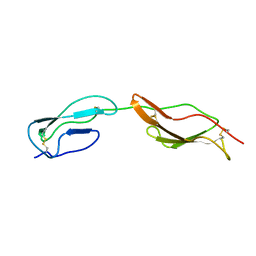 | | CR1 Sushi domains 1 and 2 | | 分子名称: | Complement receptor type 1 | | 著者 | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | 登録日 | 2013-08-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
5DRX
 
 | |
5EL1
 
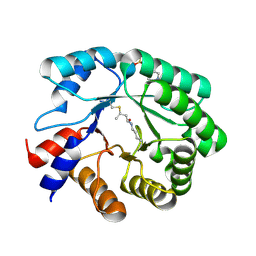 | |
5EMU
 
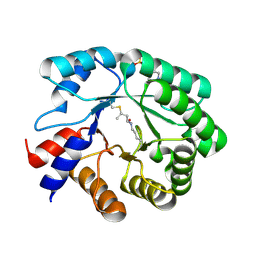 | |
5FOB
 
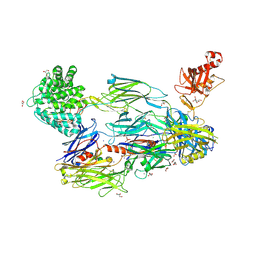 | | Crystal Structure of Human Complement C3b in complex with Smallpox Inhibitor of Complement (SPICE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COMPLEMENT C3 BETA CHAIN, ... | | 著者 | Forneris, F, Wu, J, Xue, X, Gros, P. | | 登録日 | 2015-11-18 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO8
 
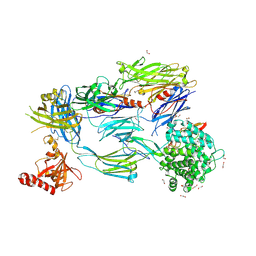 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | 著者 | Forneris, F, Wu, J, Xue, X, Gros, P. | | 登録日 | 2015-11-18 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO7
 
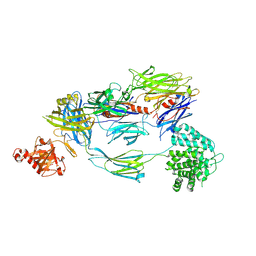 | | Crystal Structure of Human Complement C3b at 2.8 Angstrom resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN | | 著者 | Forneris, F, Wu, J, Xue, X, Gros, P. | | 登録日 | 2015-11-18 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5DRW
 
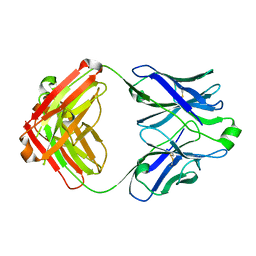 | |
5FO9
 
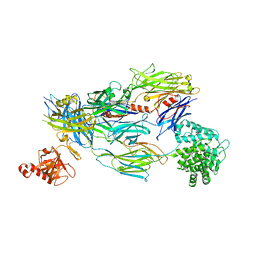 | | Crystal Structure of Human Complement C3b in Complex with CR1 (CCP15- 17) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN, ... | | 著者 | Forneris, F, Wu, J, Xue, X, Gros, P. | | 登録日 | 2015-11-18 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5EKY
 
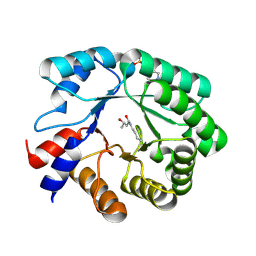 | | Crystal structure of deoxyribose-phosphate aldolase from Escherichia coli (K58E-Y96W mutant) | | 分子名称: | 1,3-BUTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Deoxyribose-phosphate aldolase | | 著者 | Classen, T, Dick, M, Pietruszka, J, Weiergraeber, O.H. | | 登録日 | 2015-11-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Mechanism-based inhibition of an aldolase at high concentrations of its natural substrate acetaldehyde: structural insights and protective strategies.
Chem Sci, 7, 2016
|
|
5FOA
 
 | | Crystal Structure of Human Complement C3b in complex with DAF (CCP2-4) | | 分子名称: | COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA CHAIN, DECAY ACCELERATING FACTOR, ... | | 著者 | Forneris, F, Wu, J, Xue, X, Gros, P. | | 登録日 | 2015-11-18 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (4.188 Å) | | 主引用文献 | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
