5IFH
 
 | | Crystal structure of the BCR Fab fragment from subset #2 case P11475 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P11475 CLL clone, Light chain of the Fab fragment from BCR derived from the P11475 CLL clone | | Authors: | Minici, C, Degano, M. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukaemia.
Nat Commun, 8, 2017
|
|
3T8I
 
 | | Structural analysis of thermostable S. solfataricus purine-specific nucleoside hydrolase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minici, C, Cacciapuoti, G, De Leo, E, Porcelli, M, Degano, M. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New Determinants in the Catalytic Mechanism of Nucleoside Hydrolases from the Structures of Two Isozymes from Sulfolobus solfataricus.
Biochemistry, 51, 2012
|
|
5DRX
 
 | |
5DRW
 
 | |
3T8J
 
 | | Structural analysis of thermostable S. solfataricus pyrimidine-specific nucleoside hydrolase | | Descriptor: | Purine nucleosidase, (IunH-1), SODIUM ION | | Authors: | Minici, C, Cacciapuoti, G, De Leo, E, Porcelli, M, Degano, M. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Determinants in the Catalytic Mechanism of Nucleoside Hydrolases from the Structures of Two Isozymes from Sulfolobus solfataricus.
Biochemistry, 51, 2012
|
|
6ZTD
 
 | | Crystal structure of the BCR Fab fragment from subset #169 case P6540 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P6540 CLL clone, Light chain of the Fab fragment from BCR derived from the P6540 CLL clone | | Authors: | Carriles, A.A, Minici, C, Degano, M. | | Deposit date: | 2020-07-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Higher-order immunoglobulin repertoire restrictions in CLL: the illustrative case of stereotyped subsets 2 and 169.
Blood, 137, 2021
|
|
3FZ0
 
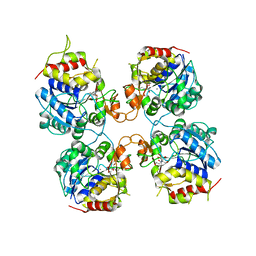 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | Deposit date: | 2009-01-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
6S81
 
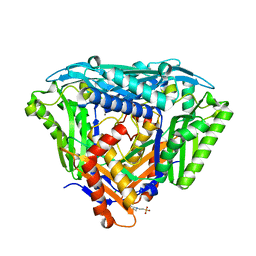 | | Crystal structure of methionine adenosyltransferase from Pyrococcus furiosus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Degano, M, Minici, C, Porcelli, M. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Structures of catalytic cycle intermediates of the Pyrococcus furiosus methionine adenosyltransferase demonstrate negative cooperativity in the archaeal orthologues.
J.Struct.Biol., 210, 2020
|
|
6S83
 
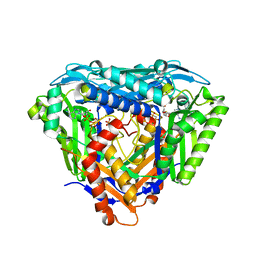 | | Crystal structure of methionine adenosyltransferase from Pyrococcus furiosus in complex with AMPPCP, SAM, and PCP | | Descriptor: | MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, PHOSPHATE ION, ... | | Authors: | Degano, M, Minici, C, Porcelli, M. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Structures of catalytic cycle intermediates of the Pyrococcus furiosus methionine adenosyltransferase demonstrate negative cooperativity in the archaeal orthologues.
J.Struct.Biol., 210, 2020
|
|
