2J5A
 
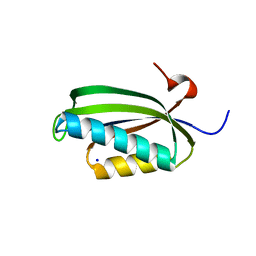 | | Folding of S6 structures with divergent amino-acid composition: pathway flexibility within partly overlapping foldons | | 分子名称: | 30S RIBOSOMAL PROTEIN S6, SODIUM ION | | 著者 | Hansson, S, Olofsson, L, Hedberg, L, Oliveberg, M, Logan, D.T. | | 登録日 | 2006-09-13 | | 公開日 | 2006-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Folding of S6 Structures with Divergent Amino Acid Composition: Pathway Flexibility within Partly Overlapping Foldons.
J.Mol.Biol., 365, 2007
|
|
1B54
 
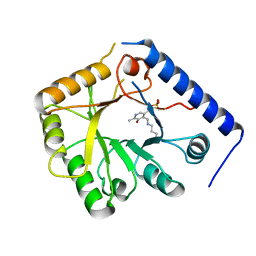 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | 著者 | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 1999-01-12 | | 公開日 | 1999-01-27 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MQ7
 
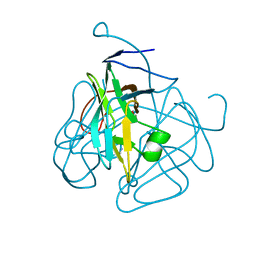 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | 著者 | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2002-09-13 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
7WB0
 
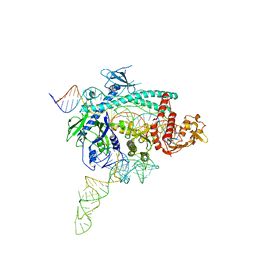 | |
7WB1
 
 | |
7WAZ
 
 | |
7WAY
 
 | |
4PG3
 
 | | Crystal structure of KRS complexed with inhibitor | | 分子名称: | LYSINE, Lysine--tRNA ligase, cladosporin | | 著者 | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.696 Å) | | 主引用文献 | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
5OWF
 
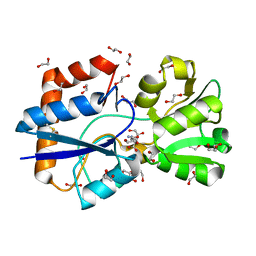 | | Structure of a LAO-binding protein mutant with glutamine | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLUTAMINE, ... | | 著者 | Shanmugaratnam, S, Banda-Vazquez, J, Sosa-Peinado, A, Hocker, B. | | 登録日 | 2017-08-31 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Redesign of LAOBP to bind novel l-amino acid ligands.
Protein Sci., 27, 2018
|
|
12E8
 
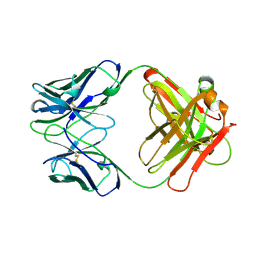 | | 2E8 FAB FRAGMENT | | 分子名称: | IGG1-KAPPA 2E8 FAB (HEAVY CHAIN), IGG1-KAPPA 2E8 FAB (LIGHT CHAIN) | | 著者 | Rupp, B, Trakhanov, S. | | 登録日 | 1998-03-14 | | 公開日 | 1998-08-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a monoclonal 2E8 Fab antibody fragment specific for the low-density lipoprotein-receptor binding region of apolipoprotein E refined at 1.9 A.
Acta Crystallogr.,Sect.D, null, 1999
|
|
4LNS
 
 | |
2RHB
 
 | | Crystal structure of Nsp15-H234A mutant- Hexamer in asymmetric unit | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Palaninathan, S, Bhardwaj, K, Alcantara, J.M.O, Guarino, L, Yi, L.L, Kao, C.C, Sacchettini, J. | | 登録日 | 2007-10-08 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15.
J.Biol.Chem., 283, 2008
|
|
1RXW
 
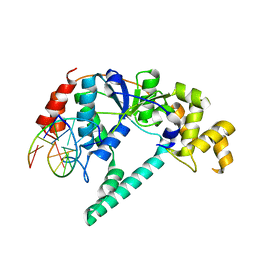 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | 分子名称: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | 著者 | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | 登録日 | 2003-12-18 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXZ
 
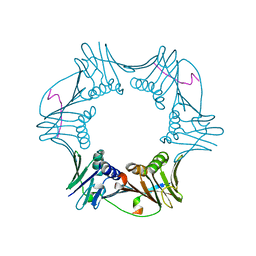 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | 分子名称: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | 著者 | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | 登録日 | 2003-12-18 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
6KKU
 
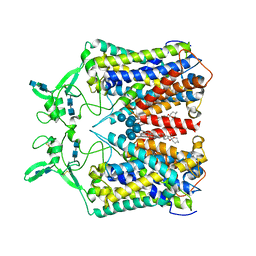 | | human KCC1 structure determined in NaCl and GDN | | 分子名称: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | 登録日 | 2019-07-27 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
7S15
 
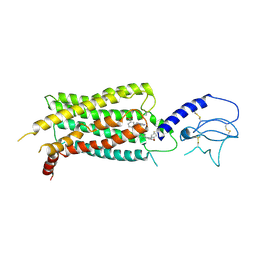 | | GLP-1 receptor bound with Pfizer small molecule agonist | | 分子名称: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | 著者 | Liu, Y, Dias, J.M, Han, S. | | 登録日 | 2021-09-01 | | 公開日 | 2022-06-08 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
1CCD
 
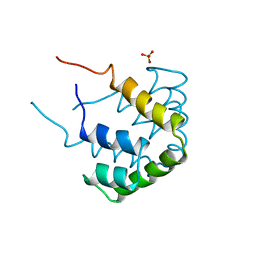 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | 分子名称: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | 著者 | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | 登録日 | 1991-09-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
7B4T
 
 | | Broadly neutralizing DARPin bnD.1 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | 分子名称: | Broadly neutralizing DARPin bnD.1, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), SULFATE ION | | 著者 | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | 登録日 | 2020-12-02 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4U
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | 分子名称: | Broadly neutralizing DARPin bnD.2, CALCIUM ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | 著者 | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | 登録日 | 2020-12-02 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4V
 
 | | Broadly neutralizing DARPin bnD.2 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | 分子名称: | Broadly neutralizing DARPin bnD.2, CHLORIDE ION, HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505), ... | | 著者 | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | 登録日 | 2020-12-02 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
7B4W
 
 | | Broadly neutralizing DARPin bnD.3 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Broadly neutralizing DARPin bnD.3, ... | | 著者 | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | 登録日 | 2020-12-02 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
3M2T
 
 | |
3M2P
 
 | |
6KKR
 
 | | human KCC1 structure determined in KCl and detergent GDN | | 分子名称: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | 登録日 | 2019-07-27 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-08-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
6KKT
 
 | | human KCC1 structure determined in KCl and lipid nanodisc | | 分子名称: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | 登録日 | 2019-07-27 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-08-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
