5HOR
 
 | | Crystal structure of c-Met-M1250T in complex with SAR125844. | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Houtmann, J, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HLW
 
 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | 分子名称: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | 登録日 | 2016-01-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOA
 
 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | 分子名称: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | 著者 | Vallee, F, Marquette, J.-P. | | 登録日 | 2016-01-19 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
7ZT6
 
 | |
7ZVT
 
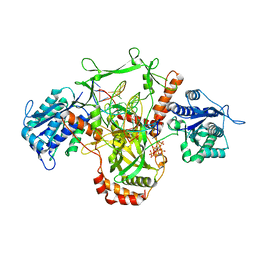 | | CryoEM structure of Ku heterodimer bound to DNA | | 分子名称: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | 登録日 | 2022-05-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
4F0I
 
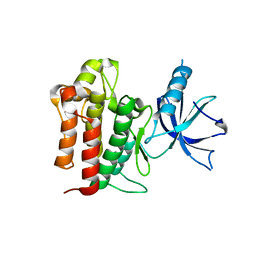 | | Crystal structure of apo TrkA | | 分子名称: | High affinity nerve growth factor receptor | | 著者 | Liu, J. | | 登録日 | 2012-05-04 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | The Crystal Structures of TrkA and TrkB Suggest Key Regions for Achieving Selective Inhibition.
J.Mol.Biol., 423, 2012
|
|
7Z6O
 
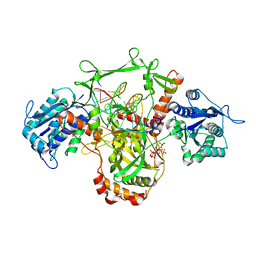 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | 分子名称: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Varela, P.F, Charbonnier, J.B. | | 登録日 | 2022-03-14 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
1GYZ
 
 | |
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | 分子名称: | Transforming growth factor beta-2 proprotein | | 著者 | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | 登録日 | 2018-11-23 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
2FKX
 
 | |
6ELP
 
 | |
6EL5
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-2-FLUORO-9-PENT-4-YLNYL-9H-PURIN-6-YLAMINE, Heat shock protein HSP 90-alpha | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-27 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EI5
 
 | |
6ELO
 
 | |
6ELN
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Heat shock protein HSP 90-alpha, SULFATE ION | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-09-29 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EY9
 
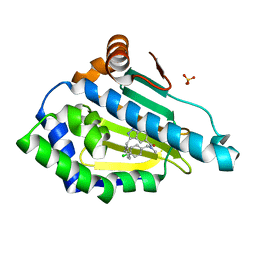 | |
6EY8
 
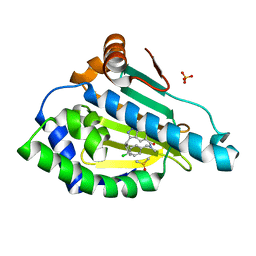 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha, SULFATE ION, ... | | 著者 | Musil, D, Lehmann, M, Buchstaller, H.-P. | | 登録日 | 2017-11-11 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EYA
 
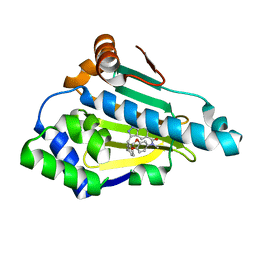 | |
6EYB
 
 | |
6F1N
 
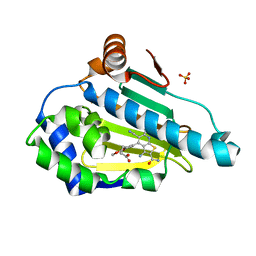 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | 分子名称: | 4-[5-[2-aminocarbonyl-3,6-bis(azanyl)-5-cyano-thieno[2,3-b]pyridin-4-yl]-2-methoxy-phenoxy]butanoic acid, Heat shock protein HSP 90-alpha, SULFATE ION | | 著者 | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | 登録日 | 2017-11-22 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6YVE
 
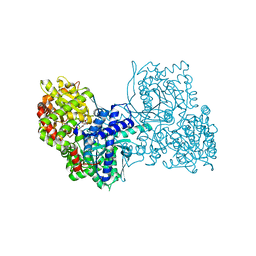 | | Glycogen phosphorylase b in complex with pelargonidin 3-O-beta-D-glucoside | | 分子名称: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | 著者 | Drakou, C.E, Gardeli, C, Tsialtas, I, Alexopoulos, S, Mallouchos, A, Koulas, S, Tsagkarakou, A, Asimakopoulos, D, Leonidas, D.D, Psarra, A.M, Skamnaki, V.T. | | 登録日 | 2020-04-28 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Affinity Crystallography Reveals Binding of Pomegranate Juice Anthocyanins at the Inhibitor Site of Glycogen Phosphorylase: The Contribution of a Sugar Moiety to Potency and Its Implications to the Binding Mode.
J.Agric.Food Chem., 68, 2020
|
|
6QC0
 
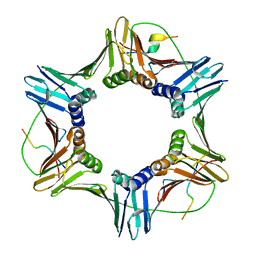 | |
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-12-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-12-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-12-23 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
