6LFK
 
 | |
1DJM
 
 | | SOLUTION STRUCTURE OF BEF3-ACTIVATED CHEY FROM ESCHERICHIA COLI | | Descriptor: | CHEMOTAXIS PROTEIN Y | | Authors: | Cho, H.S, Lee, S.Y, Yan, D, Pan, X, Parkinson, J.S, Kustu, S, Wemmer, D.E, Pelton, J.G. | | Deposit date: | 1999-12-03 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of activated CheY.
J.Mol.Biol., 297, 2000
|
|
6ISE
 
 | | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of AMPPNP bound CK2 alpha from C. neoformans
To be published
|
|
6ISJ
 
 | | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Cho, H.S. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CX-4945 bound CK2 alpha from C. neoformans
To be published
|
|
6K74
 
 | |
6K77
 
 | |
7COX
 
 | |
8OHM
 
 | |
6LBX
 
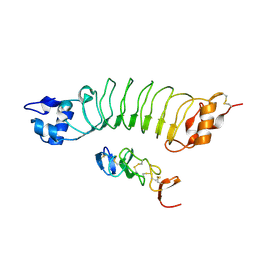 | | Crystal structure of HER2 Domain IV and Rb-H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2, Repebody (Rb-H2) | | Authors: | Cho, H.S, Cha, J.S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Computationally-guided design and affinity improvement of a protein binder targeting a specific site on HER2
Comput Struct Biotechnol J, 19, 2021
|
|
6KO6
 
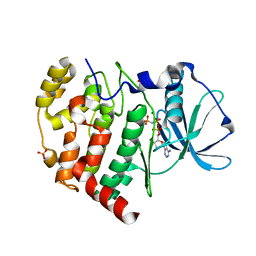 | | Crystal structure of AMPPNP bound Cka1 from C. neoformans | | Descriptor: | CMGC/CK2 protein kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
6K3L
 
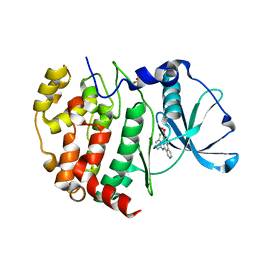 | | Crystal structure of CX-4945 bound Cka1 from C. neoformans | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CMGC/CK2 protein kinase, SULFATE ION | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
8GTG
 
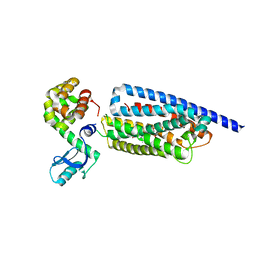 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTI
 
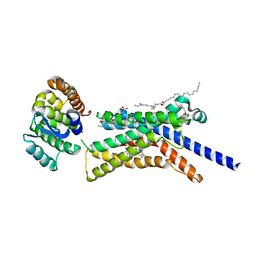 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
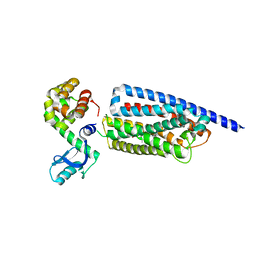 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
7E5B
 
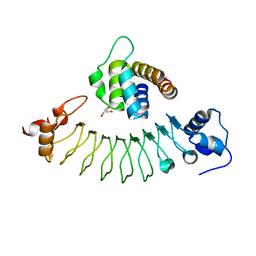 | | Crystal structure of ASC PYD Domain and Rb-B7 | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, GLYCEROL, Repebody (Rb-B7) | | Authors: | Cho, H.S, Cha, J.S. | | Deposit date: | 2021-02-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Oligomeric states of ASC specks regulate inflammatory responses by inflammasome in the extracellular space.
Cell Death Discov, 9, 2023
|
|
4UV7
 
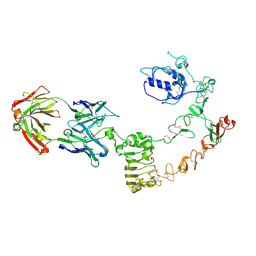 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
1KRX
 
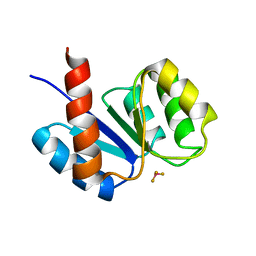 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
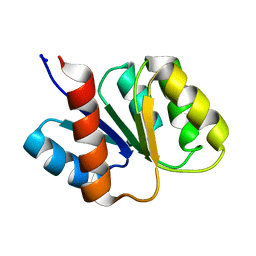 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
4D3C
 
 | |
5XHP
 
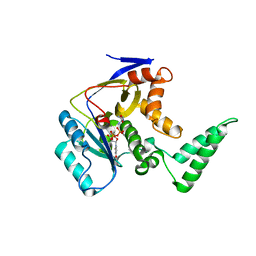 | | Transferase with ligands | | Descriptor: | ARGININE, MANGANESE (II) ION, Putative cytoplasmic protein, ... | | Authors: | Park, J, Yoo, Y, Kim, Y.H, Cho, H.S. | | Deposit date: | 2017-04-22 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of L-arginine and UDP bounded glycosyltransfease
To Be Published
|
|
9AZ7
 
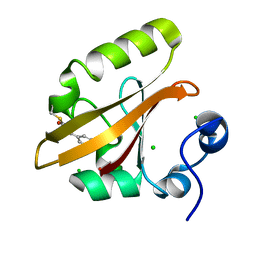 | | Chloride Sites in Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, CHLORIDE ION, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
9AZ9
 
 | | Chloride Sites in Photoactive Yellow Protein (Chloride-Free Reference Structure) | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | Deposit date: | 2024-03-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
1J56
 
 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
