3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
3Q6Z
 
 | | HUman PARP14 (ARTD8)-Macro domain 1 in complex with adenosine-5-diphosphoribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3Q71
 
 | | Human parp14 (artd8) - macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sehic, A, Thorsell, A.G, Tresaugues, L, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
4GW8
 
 | | Human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and Leucettine L41 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1,3-benzodioxol-5-ylmethyl)-2-(phenylamino)-4H-imidazol-4-one, Consensus peptide (Pimtide), ... | | Authors: | Filippakopoulos, P, Bullock, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-01 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity, cocrystal structures, and neuroprotective properties of leucettines, a family of protein kinase inhibitors derived from the marine sponge alkaloid leucettamine B.
J.Med.Chem., 55, 2012
|
|
4Z3C
 
 | | Zinc finger region of human TET3 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), Methylcytosine dioxygenase, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Dong, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-31 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 2017
|
|
4Z6H
 
 | | Crystal structure of BRD9 bromodomain in complex with a valerolactam quinolone ligand | | Descriptor: | 1,4-dimethyl-7-(2-oxopiperidin-1-yl)quinolin-2(1H)-one, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6FT8
 
 | | Crystal structure of CLK1 in complex with inhibitor 8g | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)-1~{H}-pyrrolo[3,4-g]indol-8-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT9
 
 | | Crystal structure of CLK1 in complex with inhibitor 16 | | Descriptor: | 2-bromanyl-3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, BROMIDE ION, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
5ACB
 
 | | Crystal Structure of the Human Cdk12-Cyclink Complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Mackenzie, A, Goubin, S, Strain-Damerell, C, Mahajan, P, Tallant, C, Chalk, R, Wiggers, H, Kopec, J, Fitzpatrick, F, Burgess-Brown, N, Carpenter, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Targeting of Remote Cysteine Residues to Develop Cdk12 and Cdk13 Inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
6G34
 
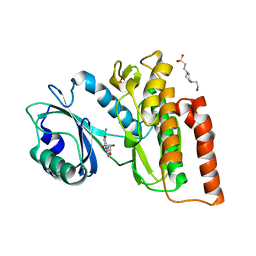 | | Crystal structure of haspin in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G38
 
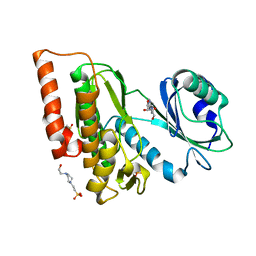 | | Crystal structure of haspin in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G37
 
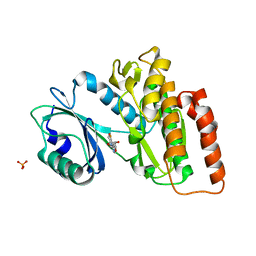 | | Crystal structure of haspin in complex with 5-fluorotubercidin | | Descriptor: | 5-fluorotubercidin, PHOSPHATE ION, Serine/threonine-protein kinase haspin | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G3A
 
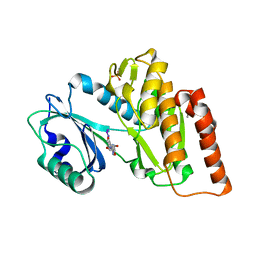 | | Crystal structure of haspin F605T mutant in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4Z6I
 
 | | Crystal structure of BRD9 bromodomain in complex with a substituted valerolactam quinolone ligand | | Descriptor: | Bromodomain-containing protein 9, tert-butyl [(2R,3S)-1-(1,4-dimethyl-2-oxo-1,2-dihydroquinolin-7-yl)-6-oxo-2-phenylpiperidin-3-yl]carbamate | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Newman, J.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-04-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AIL
 
 | | Human PARP9 2nd macrodomain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POLY [ADP-RIBOSE] POLYMERASE 9, ... | | Authors: | Sieg, C, Shrestha, L, Talon, R, Sorrell, F, Williams, E, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | Deposit date: | 2015-02-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Parp9 2Nd Macrodomain
To be Published
|
|
6G33
 
 | | Crystal structure of CLK1 in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Dual specificity protein kinase CLK1, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G35
 
 | | Crystal structure of haspin in complex with 5-bromotubercidin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-bromotubercidin, BROMIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G39
 
 | | Crystal structure of haspin F605Y mutant in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4ZQL
 
 | | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor | | Descriptor: | 3,4-dimethoxy-N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, DIMETHYL SULFOXIDE, PENTAETHYLENE GLYCOL, ... | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor
To Be Published
|
|
6G36
 
 | | Crystal structure of haspin in complex with 5-chlorotubercidin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chlorotubercidin, COBALT (II) ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6GY5
 
 | | Crystal structure of the kelch domain of human KLHL20 in complex with DAPK1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Chen, Z, Hozjan, V, Strain-Damerell, C, Williams, E, Wang, D, Cooper, C.D.O, Sanvitale, C.E, Fairhead, M, Carpenter, E.P, Pike, A.C.W, Krojer, T, Srikannathasan, V, Sorrell, F, Johansson, C, Mathea, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase.
Structure, 27, 2019
|
|
6GPK
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (E157Q) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GLYCEROL, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
4FU6
 
 | | Crystal structure of the PSIP1 PWWP domain | | Descriptor: | GLYCEROL, PC4 and SFRS1-interacting protein, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Xu, C, Wu, H, Dong, A, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the nucleosome-binding PWWP domain.
Trends Biochem.Sci., 39, 2014
|
|
