3T4G
 
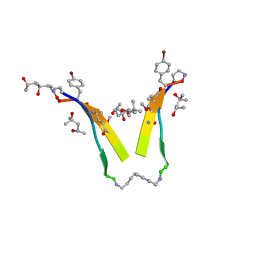 | | AIIGLMV segment from Alzheimer's Amyloid-Beta displayed on 54-membered macrocycle scaffold | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cyclic pseudo-peptide (ORN)AIIGLMV(ORN)KF(HAO)(4BF)K | | Authors: | Zhao, M, Liu, C, Cheng, P.N, Eisenberg, D, Nowick, J.S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amyloid beta-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity.
Nat Chem, 4, 2012
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
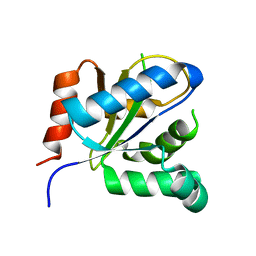
3VBC
 
 | |
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
2KNO
 
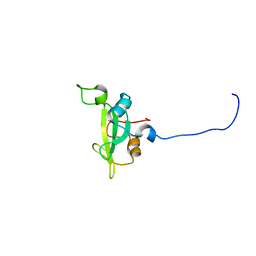 | |
2OTA
 
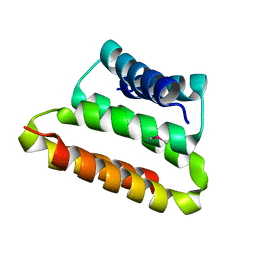 | | Crystal structure of the UPF0352 protein CPS_2611 from Colwellia psychrerythraea. NESG target CsR4. | | Descriptor: | UPF0352 protein CPS_2611 | | Authors: | Vorobiev, S.M, Zhou, W, Su, M, Seetharaman, J, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-07 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the UPF0352 protein CPS_2611 from Colwellia psychrerythraea. NESG target CsR4.
To be Published
|
|
2KKM
 
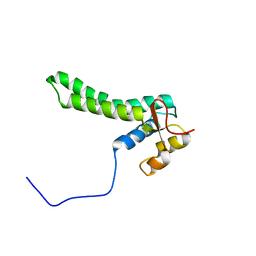 | | Solution NMR structure of yeast protein YOR252W [residues 38-178]: Northeast Structural Genomics Consortium target YT654 | | Descriptor: | Translation machinery-associated protein 16 | | Authors: | Cort, J.R, Yee, A, Liu, C, Arrowsmith, C.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast protein YOR252W
To be Published
|
|
8IKS
 
 | |
8IK7
 
 | |
8IKP
 
 | |
8IKB
 
 | |
8IY7
 
 | |
8IZZ
 
 | |
4P32
 
 | | Crystal structure of E. coli LptB in complex with ADP-magnesium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P33
 
 | | Crystal structure of E. coli LptB-E163Q in complex with ATP-sodium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P31
 
 | | Crystal structure of a selenomethionine derivative of E. coli LptB in complex with ADP-Magensium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, MAGNESIUM ION | | Authors: | Sherman, D.J, Lazarus, M.B, Murphy, L, Liu, C, Walker, S, Ruiz, N, Kahne, D. | | Deposit date: | 2014-03-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MYD
 
 | | 1.37 Angstrom Crystal Structure of E. Coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) in complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-27 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
