1LEI
 
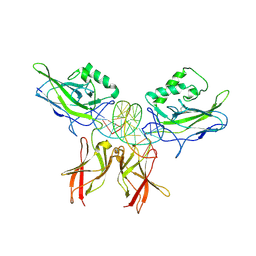 | | The kB DNA sequence from the HLV-LTR functions as an allosteric regulator of HIV transcription | | Descriptor: | 5'-D(*CP*TP*CP*AP*GP*GP*GP*AP*AP*AP*GP*TP*AP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*5ITP*AP*CP*5ITP*5ITP*5ITP*CP*CP*CP*TP*GP*AP*G)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Chen-Park, F, Huang, D.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The kB DNA sequence from the HIV Long Terminal Repeat functions as an allosteric regulator of HIV transcription
J.Biol.Chem., 277, 2002
|
|
8URP
 
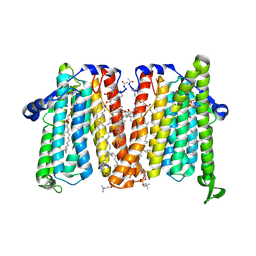 | | Cholinephosphotransferase in complex with CDP-choline and phosphatidylcholine | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cholinephosphotransferase 1, ... | | Authors: | Roberts, J.R, Maeda, S, Ohi, M.D. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for catalysis and selectivity of phospholipid synthesis by eukaryotic choline-phosphotransferase.
Nat Commun, 16, 2025
|
|
8URT
 
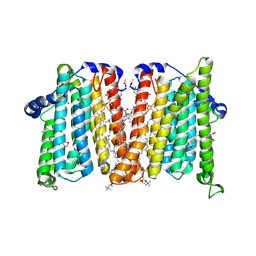 | | Cholinephosphotransferase in complex with selective inhibitor chelerythrine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Cholinephosphotransferase 1, ... | | Authors: | Roberts, J.R, Maeda, S, Ohi, M.D. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-30 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and selectivity of phospholipid synthesis by eukaryotic choline-phosphotransferase.
Nat Commun, 16, 2025
|
|
7ZMZ
 
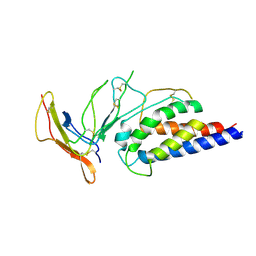 | | Engineered Interleukin 2 bound to CD25 receptor | | Descriptor: | Interleukin-2, Interleukin-2 receptor subunit alpha | | Authors: | Fyfe, P.K, Moraga, I, Gaggero, S, Mitra, S. | | Deposit date: | 2022-04-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | IL-2 is inactivated by the acidic pH environment of tumors enabling engineering of a pH-selective mutein.
Sci Immunol, 7, 2022
|
|
3SIT
 
 | |
6HX1
 
 | | IRE1 ALPHA IN COMPLEX WITH imidazo[1,2-b]pyridazin-8-amine compound 2 | | Descriptor: | 6-chloranyl-~{N}-(cyclopropylmethyl)-3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Augustin, M.A, Krapp, S, Bayliss, R, Collins, I. | | Deposit date: | 2018-10-15 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
6AMS
 
 | |
6AMQ
 
 | |
6Y6F
 
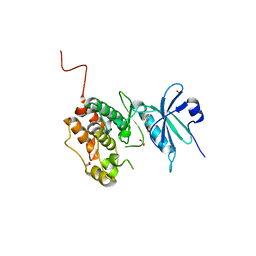 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6Y6H
 
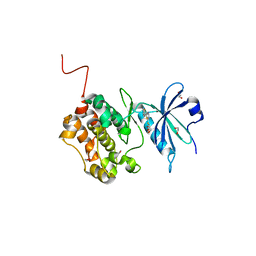 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6XVF
 
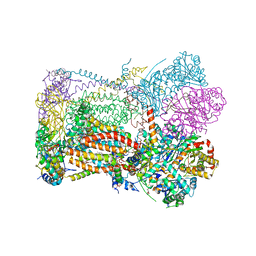 | | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites.
Front Cell Infect Microbiol, 10, 2020
|
|
6HV0
 
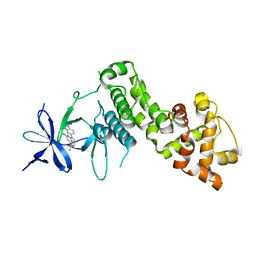 | | IRE1 kinase/RNase in complex with imidazo[1,2-b]pyridazin-8-amine compound 33 | | Descriptor: | 6-chloranyl-3-(2~{H}-indazol-5-yl)-~{N}-propan-2-yl-imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Bayliss, R, Bhatia, C, Collins, I. | | Deposit date: | 2018-10-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
6AP4
 
 | |
6PMO
 
 | |
6POM
 
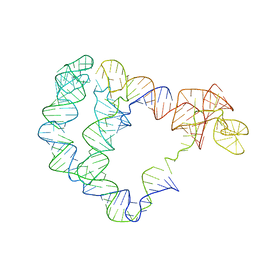 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OL3
 
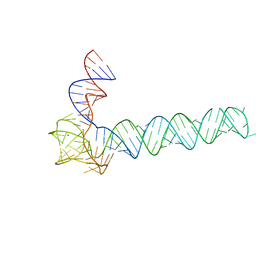 | | Crystal structure of an adenovirus virus-associated RNA | | Descriptor: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | Authors: | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
4ZNN
 
 | | MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56 | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.41 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RIK
 
 | | Amyloid forming segment, AVVTGVTAV, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 69-77 | | Descriptor: | Alpha-synuclein | | Authors: | Guenther, E.L, Sawaya, M.R, Ivanova, M, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
4RIL
 
 | | Structure of the amyloid forming segment, GAVVTGVTAVA, from the NAC domain of Parkinson's disease protein alpha-synuclein, residues 68-78, determined by electron diffraction | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2014-10-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.43 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
6SSH
 
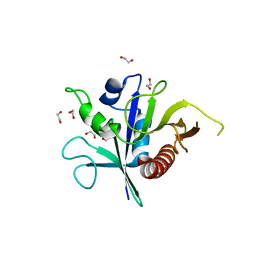 | | Structure of the TSC2 GAP domain | | Descriptor: | 1,2-ETHANEDIOL, GTPase activator-like protein | | Authors: | Hansmann, P, Kiontke, S, Kummel, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the TSC2 GAP Domain: Mechanistic Insight into Catalysis and Pathogenic Mutations.
Structure, 28, 2020
|
|
4QIO
 
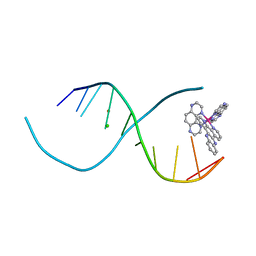 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCGCCIA) at high resolution | | Descriptor: | 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*IP*A)-3', BARIUM ION, CHLORIDE ION, ... | | Authors: | Gurung, S.P, Hall, J.P, Cardin, C.J. | | Deposit date: | 2014-06-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Inosine Can Increase DNA's Susceptibility to Photo-oxidation by a Ru II Complex due to Structural Change in the Minor Groove.
Chemistry, 23, 2017
|
|
4R8J
 
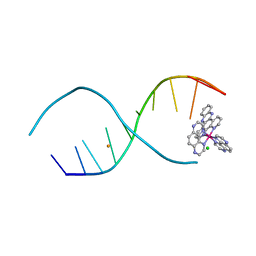 | | d(TCGGCGCCGA) with lambda-[Ru(TAP)2(dppz)]2+ soaked in D2O | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*(THM)P*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Gurung, S.P, Winter, G.W, Cardin, C.J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Monitoring one-electron photo-oxidation of guanine in DNA crystals using ultrafast infrared spectroscopy.
Nat Chem, 7, 2015
|
|
3FO4
 
 | |
7KEF
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
