3EQB
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5Z1W
 
 | |
3DY7
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | (5S)-4,5-difluoro-6-[(2-fluoro-4-iodophenyl)imino]-N-(2-hydroxyethoxy)cyclohexa-1,3-diene-1-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-07-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8J2X
 
 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
6VQL
 
 | |
3DNG
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | (5S)-5-(2-amino-2-oxoethyl)-4-oxo-N-[(3-oxo-3,4-dihydro-2H-1,4-benzoxazin-6-yl)methyl]-3,4,5,6,7,8-hexahydro[1]benzothieno[2,3-d]pyrimidine-2-carboxamide, CALCIUM ION, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
4KXF
 
 | |
3DPF
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
3DPE
 
 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | Descriptor: | CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, Neutrophil collagenase, ... | | Authors: | Pochetti, G, Montanari, R, Mazza, F. | | Deposit date: | 2008-07-08 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
8J2W
 
 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2Y
 
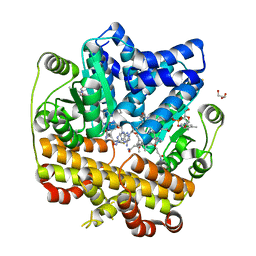 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
3JCO
 
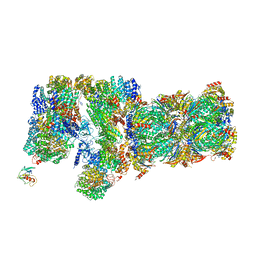 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LXY
 
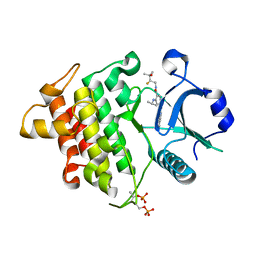 | | IRAK4 in complex with inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-fluoranyl-3-methyl-3-oxidanyl-butyl]-6-[(6-fluoranylpyrazolo[1,5-a]pyrimidin-5-yl)amino]-4-(propan-2-ylamino)pyridine-3-carboxamide, SULFATE ION | | Authors: | Ghosh, K, Bose, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-11-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of Nicotinamides as Potent and Selective IRAK4 Inhibitors with Efficacy in a Murine Model of Psoriasis.
Acs Med.Chem.Lett., 11, 2020
|
|
6AZW
 
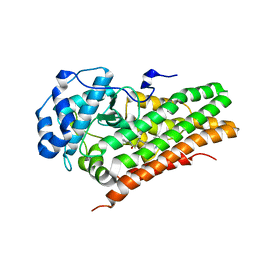 | | IDO1/FXB-001116 crystal structure | | Descriptor: | (2R)-N-(4-cyanophenyl)-2-[cis-4-(quinolin-4-yl)cyclohexyl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A, Lammens, A, Steinbacher, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CNB
 
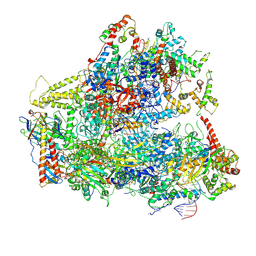 | | Yeast RNA polymerase III initial transcribing complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CND
 
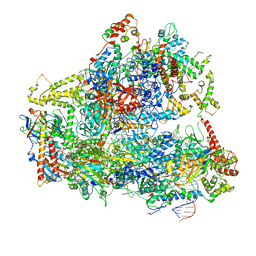 | | Yeast RNA polymerase III natural open complex (nOC) | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNC
 
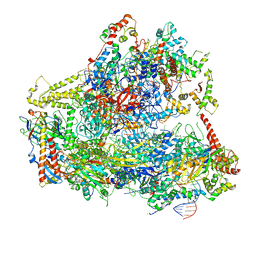 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNF
 
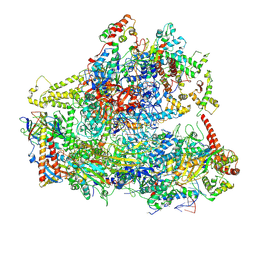 | | Yeast RNA polymerase III elongation complex | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6AZV
 
 | | IDO1/BMS-978587 crystal structure | | Descriptor: | (1R,2S)-2-(4-[bis(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6LR4
 
 | | Molecular basis for inhibition of human gamma-secretase by small molecule | | Descriptor: | (2S)-2-hydroxy-3-methyl-N-[(2S)-1-[[(5S)-3-methyl-4-oxo-2,5-dihydro-1H-3-benzazepin-5-yl]amino]-1-oxopropan-2-yl]butanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
6LQG
 
 | | Human gamma-secretase in complex with small molecule Avagacestat | | Descriptor: | (2R)-2-[(4-chlorophenyl)sulfonyl-[[2-fluoranyl-4-(1,2,4-oxadiazol-3-yl)phenyl]methyl]amino]-5,5,5-tris(fluoranyl)pentanamide, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Guo, X, Lei, J, Shi, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2021-01-27 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of gamma-secretase inhibition and modulation by small molecule drugs.
Cell, 184, 2021
|
|
7E26
 
 | | Structure of PfFNT in apo state | | Descriptor: | Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | Deposit date: | 2021-02-04 | | Release date: | 2021-08-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
5W65
 
 | | RNA polymerase I Initial Transcribing Complex State 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
 | | RNA Polymerase I Initial Transcribing Complex State 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
