5CKA
 
 | | Human beta-2 microglobulin double mutant W60G-N83V | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CKG
 
 | | Human beta-2 microglobulin mutant V85E | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL | | Authors: | Sala, B.M, De Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-15 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability.
Sci Rep, 6, 2016
|
|
5CFH
 
 | |
6F60
 
 | | The x-ray structure of bovine pancreatic ribonuclease in complex with a five-coordinate platinum(II) compound containing a sugar ligand | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, five-coordinate platinum(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2017-12-04 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Five-Coordinate Platinum(II) Compounds Containing Sugar Ligands: Synthesis, Characterization, Cytotoxic Activity, and Interaction with Biological Macromolecules.
Inorg Chem, 57, 2018
|
|
5C26
 
 | | Crystal structure of SYK in complex with compound 1 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}benzamide, GLU-VAL-PTR-GLU-SER-PRO, Tyrosine-protein kinase SYK | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
5C27
 
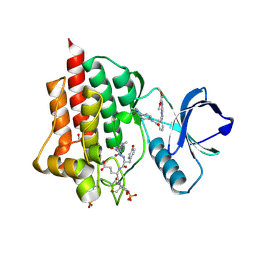 | | Crystal structure of SYK in complex with compound 2 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
5JQ0
 
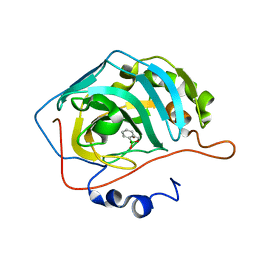 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH=8.7 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
5JQT
 
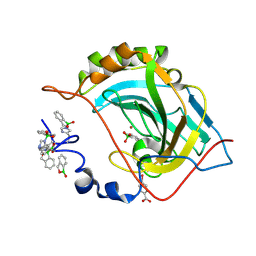 | | Crystal structure of human carbonic anhydrase II in complex with Benzoxaborole at pH 7.4 | | Descriptor: | 1,1-dihydroxy-1,3-dihydro-2,1-benzoxaborol-1-ium, 2,1-benzoxaborol-1(3H)-ol, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | Alterio, V, Esposito, D, Di Fiore, A, De Simone, G. | | Deposit date: | 2016-05-05 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Benzoxaborole as a new chemotype for carbonic anhydrase inhibition.
Chem.Commun.(Camb.), 52, 2016
|
|
8PMA
 
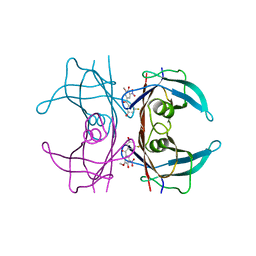 | | Crystal structure of human V30M transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PMO
 
 | | Crystal structure of human V122I transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PM8
 
 | | V30M Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
8PM9
 
 | | Crystal structure of human wild type transthyretin in complex with PITB (Pharmacokinetically Improved TTR Binder) | | Descriptor: | (3-fluoranyl-5-oxidanyl-phenyl)-(3-methoxy-5-nitro-4-oxidanyl-phenyl)methanone, Transthyretin | | Authors: | Varejao, N, Pinheiro, F, Pallares, I, Ventura, S, Reverter, D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PITB: A high affinity transthyretin aggregation inhibitor with optimal pharmacokinetic properties.
Eur.J.Med.Chem., 261, 2023
|
|
1E6U
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-08-23 | | Release date: | 2000-10-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
5AFO
 
 | |
1E7S
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase K140R | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Zuccotti, S, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1EBT
 
 | |
1E7R
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1E7Q
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase S107A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1EBC
 
 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
6XAT
 
 | | Crystal structure of the human FoxP4 DNA binding Domain | | Descriptor: | FOXP4 protein, SODIUM ION | | Authors: | VIllalobos, P, Castro-Fernandez, V, Medina, E, Gonzalez-Ordenes, F, Maturana, P, Herrera-Morande, A, Ramirez-Sarmiento, C.A, Babul, J. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unraveling the folding and dimerization properties of the human FoxP subfamily of transcription factors.
Febs Lett., 597, 2023
|
|
6XIO
 
 | | ADP-dependent kinase complex with fructose-6-phosphate and ADPbetaS | | Descriptor: | 5'-O-[(R)-HYDROXY(THIOPHOSPHONOOXY)PHOSPHORYL]ADENOSINE, 6-O-phosphono-beta-D-fructofuranose, ADP-dependent phosphofructokinase, ... | | Authors: | Munoz, S, Gonzalez-Ordenes, F, Fuentes, N, Maturana, P, Herrera-Morande, A, Villalobos, P, Castro-Fernandez, V. | | Deposit date: | 2020-06-20 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of an ancestral ADP-dependent kinase with fructose-6P reveals key residues for binding, catalysis, and ligand-induced conformational changes.
J.Biol.Chem., 296, 2020
|
|
6XEQ
 
 | | Crystal structure of the tetrameric 6-phosphogluconate dehydrogenase from Gluconobacter oxidans | | Descriptor: | 6-phosphogluconate dehydrogenase, SULFATE ION | | Authors: | Maturana, P, Roversi, P, Castro-Fernandez, V, Herrera-Morande, A, Garratt, R.C, Cabrera, R. | | Deposit date: | 2020-06-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the 6-phosphogluconate dehydrogenase from Gluconobacter oxydans reveals tetrameric 6PGDHs as the crucial intermediate in the evolution of structure and cofactor preference in the 6PGDH family [version 1; peer review: 1 approved, 1 approved with reservations]
Wellcome Open Res, 6, 2021
|
|
6P5L
 
 | | Crystal Structure of Ubl123 with an EZH2 peptide | | Descriptor: | PRO-ARG-LYS-LYS-LYS-ARG-LYS-HIS, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural Basis of the Interaction Between Ubiquitin Specific Protease 7 and Enhancer of Zeste Homolog 2.
J.Mol.Biol., 432, 2020
|
|
6VMJ
 
 | |
7Z74
 
 | | PI3KC2a core in complex with PITCOIN2 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ~{N}-[4-(3-hydroxyphenyl)-1,3-thiazol-2-yl]-2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-ethanamide | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
