5LAR
 
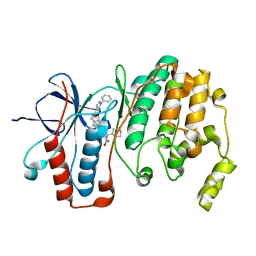 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
5LVQ
 
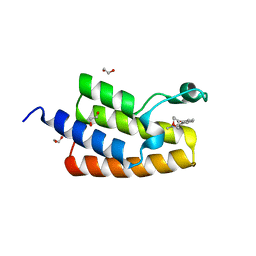 | | Crystal structure of human PCAF bromodomain in complex with compound-D (CPD-D), N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5LWM
 
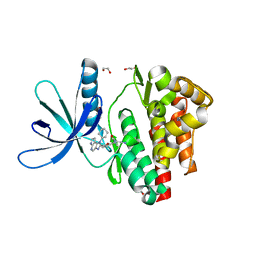 | | Crystal structure of JAK3 in complex with Compound 4 (FM381) | | Descriptor: | 1,2-ETHANEDIOL, 1-phenylurea, 2-cyano-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl)furan-2-yl]-~{N},~{N}-dimethyl-prop-2-enamide, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
6I5K
 
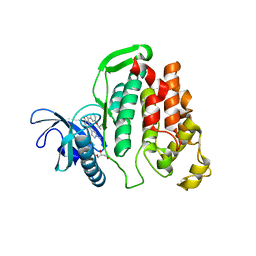 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN345 (derivative of compound 12h) | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-(3-propan-2-yloxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
3MY0
 
 | | Crystal structure of the ACVRL1 (ALK1) kinase domain bound to LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Serine/threonine-protein kinase receptor R3 | | Authors: | Chaikuad, A, Alfano, I, Cooper, C, Mahajan, P, Daga, N, Sanvitale, C, Fedorov, O, Petrie, K, Savitsky, P, Gileadi, O, Sethi, R, Krojer, T, Muniz, J.R.C, Pike, A.C.W, Vollmar, M, Carpenter, C.P, Ugochukwu, E, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-08 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A small molecule targeting ALK1 prevents Notch cooperativity and inhibits functional angiogenesis.
Angiogenesis, 18, 2015
|
|
3MTF
 
 | | Crystal structure of the ACVR1 kinase in complex with a 2-aminopyridine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1, ... | | Authors: | Chaikuad, A, Sanvitale, C, Cooper, C, Mahajan, P, Daga, N, Petrie, K, Alfano, I, Canning, P, Krojer, T, Vollmar, M, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A new class of small molecule inhibitor of BMP signaling.
Plos One, 8, 2013
|
|
5LVR
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-E (CPD-E) | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-phenyl-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5LWN
 
 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | Descriptor: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | Authors: | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-18 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
4LOP
 
 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6I5L
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN316 (derivative of compound 12h) | | Descriptor: | 3-(3-cyclobutylphenyl)-5-(1-methylpyrazol-4-yl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6I5I
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7OAJ
 
 | | Crystal structure of pseudokinase CASK in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(cyclopentylamino)-2-[(3,4-dichlorophenyl)methylamino]-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAK
 
 | | Crystal structure of pseudokinase CASK in complex with compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAI
 
 | | Crystal structure of pseudokinase CASK in complex with PFE-PKIS12 | | Descriptor: | 1,2-ETHANEDIOL, 4-(Cyclopentylamino)-2-[(2,5-dichlorophenyl)methylamino]-N-[3-(2-oxo-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAL
 
 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7ORF
 
 | | Crystal structure of JNK3 in complex with FMU-001-367 (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 10, ... | | Authors: | Chaikuad, A, Koch, P, Laufer, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OPS
 
 | | Crystal structure of haspin in complex with ZW282 (compound 2a) | | Descriptor: | 2-methylsulfanyl-10-nitro-pyrido[3,4-g]quinazoline, GLYCEROL, Serine/threonine-protein kinase haspin | | Authors: | Chaikuad, A, Anizon, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-01 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synthesis and biological evaluation of Haspin inhibitors: Kinase inhibitory potency and cellular activity.
Eur.J.Med.Chem., 236, 2022
|
|
7ORE
 
 | | Crystal structure of JNK3 in complex with light-activated covalent inhibitor MR-II-249 with both non-covalent and covalent binding modes (compound 4) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[(5Z)-9-[[4-[5-(4-fluorophenyl)-3-methyl-2-methylsulfanyl-imidazol-4-yl]pyridin-2-yl]amino]-11,12-dihydrobenzo[c][1,2]benzodiazocin-2-yl]butanamide, Mitogen-activated protein kinase 10 | | Authors: | Chaikuad, A, Reynders, M, Trauner, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-05 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Controlling the Covalent Reactivity of a Kinase Inhibitor with Light.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YZ4
 
 | | Crystal structure of MKK7 (MAP2K7) with ibrutinib bound at allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-06 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6FT8
 
 | | Crystal structure of CLK1 in complex with inhibitor 8g | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)-1~{H}-pyrrolo[3,4-g]indol-8-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT9
 
 | | Crystal structure of CLK1 in complex with inhibitor 16 | | Descriptor: | 2-bromanyl-3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, BROMIDE ION, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
3KHU
 
 | | Crystal structure of human UDP-glucose dehydrogenase Glu161Gln, in complex with thiohemiacetal intermediate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, UDP-glucose 6-dehydrogenase, ... | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Guo, K, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Evidence That Catalytic Reaction of Human UDP-glucose 6-Dehydrogenase Involves Covalent Thiohemiacetal and Thioester Enzyme Intermediates.
J.Biol.Chem., 287, 2012
|
|
9GFO
 
 | | iASPP-CTD fusion to p63 peptide | | Descriptor: | Tumor protein 63,RelA-associated inhibitor | | Authors: | Chaikuad, A, Akutsu, M, Lotz, R, Osterburg, C, Knapp, S, Dotsch, V. | | Deposit date: | 2024-08-12 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alternative splicing in the DBD linker region of p63 modulates binding to DNA and iASPP in vitro.
Cell Death Dis, 16, 2025
|
|
4QUT
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) complexed with Histone H4-K(ac)12 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, Histone H4, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-12 | | Release date: | 2014-07-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
J Mol Cell Biol, 8, 2016
|
|
