8OLB
 
 | | SA11 Rotavirus Non-tripsinized Triple Layered Particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Inner capsid protein VP2, ... | | Authors: | Asensio-Cob, D, Perez-Mata, C, Gomez-Blanco, J, Vargas, J, Rodriguez, J.M, Luque, D. | | Deposit date: | 2023-03-30 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of rotavirus spike proteolytic activation
To Be Published
|
|
5FWQ
 
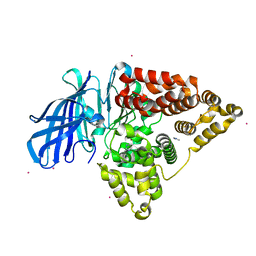 | | Apo structure of human Leukotriene A4 hydrolase | | Descriptor: | ACETATE ION, HUMAN LEUKOTRIENE A4 HYDROLASE, IMIDAZOLE, ... | | Authors: | Wittmann, S.K, Kalinowsky, L, Kramer, J, Bloecher, R, Steinhilber, D, Pogoryelov, D, Proschak, E, Heering, J. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Thermodynamic properties of leukotriene A4hydrolase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5HBH
 
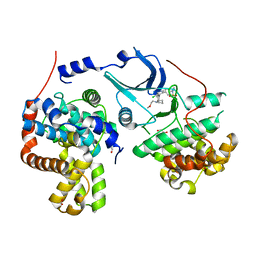 | | CDK8-CYCC IN COMPLEX WITH 5-{5-Chloro-4-[1-(2-methoxy-ethyl)-1,8-diaza-spiro[4.5]dec-8-yl]-pyridin-3-yl}-1-methyl-1,3-dihydro-benzo[c]isothiazole 2,2-dioxide | | Descriptor: | 1,2-ETHANEDIOL, 5-[5-chloranyl-4-[1-(2-methoxyethyl)-1,8-diazaspiro[4.5]decan-8-yl]pyridin-3-yl]-1-methyl-3~{H}-2,1-benzothiazole 2,2-dioxide, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
9F2Q
 
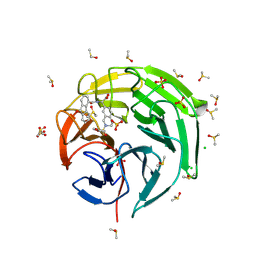 | | Crystal structure of Keap1 kelch domain in complex with a tetrahydroisoquinoline-based small molecule inhibitor at 1.2A resolution | | Descriptor: | (R)-2-(2-((4-methoxyphenyl)sulfonyl)-1,2,3,4-tetrahydroisoquinolin-7-yl)-2-(9-oxo-9H-fluorene-4-carboxamido)acetate, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Guided Conformational Restriction Leading to High-Affinity, Selective, and Cell-Active Tetrahydroisoquinoline-Based Noncovalent Keap1-Nrf2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9F2P
 
 | | Crystal structure of Keap1 kelch domain in complex with a fluorenone-based small molecule inhibitor at 1.36A resolution | | Descriptor: | (R)-2-(3-(((4-methoxyphenyl)sulfonamido)methyl)phenyl)-2-(9-oxo-9H-fluorene-4-carboxamido)acetate, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure-Guided Conformational Restriction Leading to High-Affinity, Selective, and Cell-Active Tetrahydroisoquinoline-Based Noncovalent Keap1-Nrf2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
5JBQ
 
 | |
8PIQ
 
 | | Crystal Structure of BRD4-BD1 with BI894999 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Kessler, D. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-26 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.117 Å) | | Cite: | Crystal Structure of BRD4-BD1 with BI894999
Not published
|
|
6TK3
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 30us+150us structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK6
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in neutral conditions with attached light datasets at 800fs, 2ps, 100ps, 1ns, 16ns, 1us, 30us, 150us, 1ms and 20ms | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK4
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 1ns+16ns structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
5DAA
 
 | |
8WKZ
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
6TX4
 
 | |
6TX9
 
 | |
8AJ2
 
 | |
8ARB
 
 | | Heterologous Complex of shortened Aeromonas hydrophila Type III secretion substrate AscX with Yersinia enterocolitica chaperone YscY | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AscX, Chaperone protein YscY, ... | | Authors: | Gilzer, D, Flottmann, F, Niemann, H.H. | | Deposit date: | 2022-08-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The type III secretion chaperone SctY may shield the hydrophobic export gate-binding C-terminus of its substrate SctX.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8RQA
 
 | | Crystal structure of CRBN-midi in complex with Lenalidomide | | Descriptor: | Protein cereblon, S-Lenalidomide, ZINC ION | | Authors: | Rutter, Z.J, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8ARC
 
 | |
8ADF
 
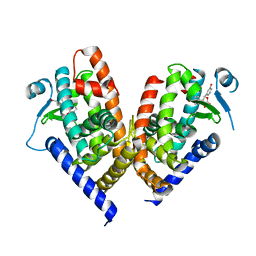 | | X-ray crystal structure of PPAR gamma ligand binding domain in complex with CZ39 | | Descriptor: | (2R)-3-(4-bromophenyl)-2-(3-hydroxyphenyl)-4-oxidanyl-2H-furan-5-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R, Pochetti, G, Meneghetti, F, Villa, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Biological Screening and Crystallographic Studies of Hydroxy gamma-Lactone Derivatives to Investigate PPAR gamma Phosphorylation Inhibition.
Biomolecules, 13, 2023
|
|
8ARA
 
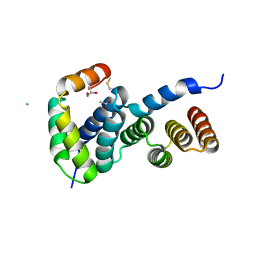 | | Heterologous Complex of Aeromonas hydrophila Type III secretion substrate AscX with Yersinia enterocolitica chaperone YscY | | Descriptor: | ACETATE ION, AscX, CHLORIDE ION, ... | | Authors: | Gilzer, D, Kowal, J.L, Niemann, H.H. | | Deposit date: | 2022-08-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The type III secretion chaperone SctY may shield the hydrophobic export gate-binding C-terminus of its substrate SctX.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5U2N
 
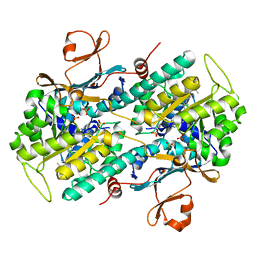 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
8RQ1
 
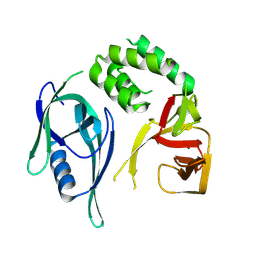 | | Crystal structure of CRBN-midi | | Descriptor: | Protein cereblon, ZINC ION | | Authors: | Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8RQ8
 
 | | Crystal structure of CRBN-midi in complex with mezigdomide | | Descriptor: | Mezigdomide, Protein cereblon, ZINC ION | | Authors: | Zollman, D, Kroupova, A, Pethe, J, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
8RQC
 
 | | Crystal structure of CRBN-midi in complex with mezigdomide and IKZF1 ZF2 | | Descriptor: | DNA-binding protein Ikaros, Mezigdomide, Protein cereblon, ... | | Authors: | Furihata, H, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
7ZYM
 
 | | Crystal Structure of EGFR-T790M/C797S in Complex with Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Niggenaber, J, Kleinboelting, S, Mueller, M.P, Rauh, D. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Addressing the Osimertinib Resistance Mutation EGFR-L858R/C797S with Reversible Aminopyrimidines.
Acs Med.Chem.Lett., 14, 2023
|
|
