8VQ3
 
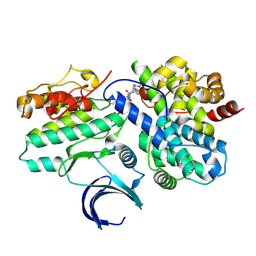 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | An allosteric cyclin E-CDK2 site mapped by paralog hopping with covalent probes.
Nat.Chem.Biol., 2024
|
|
2N98
 
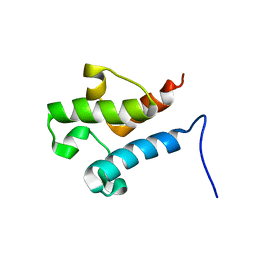 | | Solution structure of acyl carrier protein LipD from Actinoplanes friuliensis | | Descriptor: | Acyl carrier protein | | Authors: | Paul, S, Ishida, H, Liu, Z, Nguyen, L.T, Vogel, H.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of a freestanding acyl carrier protein involved in the biosynthesis of cyclic lipopeptide antibiotics.
Protein Sci., 26, 2017
|
|
5YIK
 
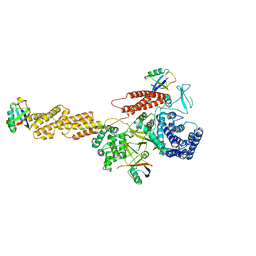 | |
4AOJ
 
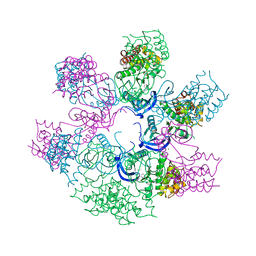 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
7F09
 
 | | Crystal structure of the HLH-Lz domain of human TFE3 | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor E3, ZINC ION | | Authors: | Yang, G, Li, P, Liu, Z, Wu, S, Zhuang, C, Qiao, H, Fang, P, Wang, J. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the dimerization mechanism of human transcription factor E3.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
3QPD
 
 | | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon | | Descriptor: | Cutinase 1 | | Authors: | Lu, A, Gosser, Y, Montclare, J.K, Liu, Z, Kong, X. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon
To be Published
|
|
3QPC
 
 | | Structure of Fusarium Solani Cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon | | Descriptor: | Cutinase | | Authors: | Lu, A, Gosser, Y, Montclare, J.K, Liu, Z, Kong, X. | | Deposit date: | 2011-02-11 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (0.978 Å) | | Cite: | Structure of Fusarium Solani Cutinase expressed in Pichia pastoris
To be Published
|
|
3R0W
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors. | | Descriptor: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2R)-1-{[(2S,3S)-5-{[(2R)-1-{[(2S)-1-amino-4-methyl-1-oxopentan-2-yl]amino}-3-chloro-1-oxopropan-2-yl]amino}-3-hydroxy-5-oxo-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | Authors: | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | Deposit date: | 2011-03-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
6BZ9
 
 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3SPK
 
 | | Tipranavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE | | Authors: | Wang, Y, Liu, Z, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2011-07-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3SO9
 
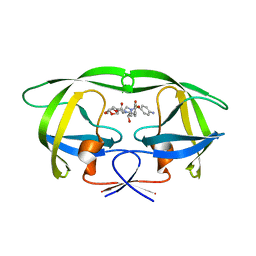 | | Darunavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Wang, Y, Liu, Z, Brunzelle, S.J, Kovari, L.C, Kovari, I.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
2JXN
 
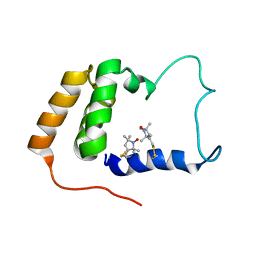 | | Solution Structure of S. cerevisiae PDCD5-like Protein Ymr074cp | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, Uncharacterized protein YMR074C | | Authors: | Hong, J, Zhang, J, Liu, Z, Shi, Y, Wu, J. | | Deposit date: | 2007-11-23 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of S. cerevisiae PDCD5-like Protein Ymr074cp Determined by Heteronuclear NMR Spectroscopy
To be Published
|
|
7D3Y
 
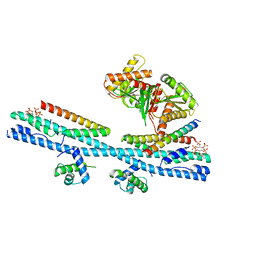 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
7D3T
 
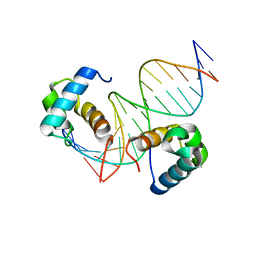 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
3QPA
 
 | |
3R0Y
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors | | Descriptor: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2S)-1-{[(2S,3S)-3-hydroxy-5-oxo-5-{[(2R)-1-oxo-3-phenyl-1-(prop-2-yn-1-ylamino)propan-2-yl]amino}-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | Authors: | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | Deposit date: | 2011-03-09 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
2LZK
 
 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2JTB
 
 | |
2MIV
 
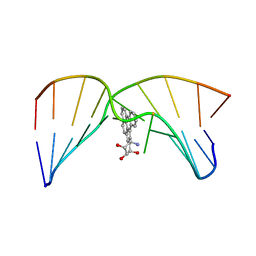 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2LX9
 
 | |
8IVB
 
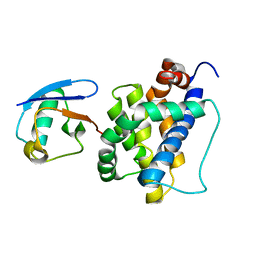 | | K113-Ubiquitinated BAK | | Descriptor: | Bcl-2 homologous antagonist/killer, Ubiquitin | | Authors: | Dong, X, Cheng, P, Hou, Y.Z, Chen, Y.K, Liu, Z. | | Deposit date: | 2023-03-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Commun Biol, 6, 2023
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6INW
 
 | | A Pericyclic Reaction enzyme | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Feng, Y, Chang, M, Wang, H, Liu, Z, Zhou, Y. | | Deposit date: | 2018-10-28 | | Release date: | 2019-07-03 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of the multifunctional SAM-dependent enzyme LepI provides insights into its catalytic mechanism.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6IJO
 
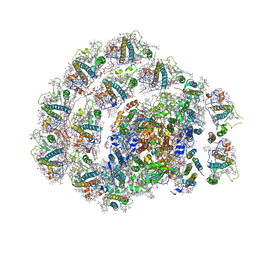 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
