6OL7
 
 | |
6MZJ
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6VRQ
 
 | | Crystal structure of gl12A21 Fab in complex with anti-idiotypic iv12 Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aljedani, S.S, Weidle, C, Pancera, M. | | Deposit date: | 2020-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Development of a VRC01-class germline targeting immunogen derived from anti-idiotypic antibodies.
Cell Rep, 35, 2021
|
|
6WAS
 
 | | Structure of D19.PA8 Fab in complex with 1FD6 16055 V1V2 scaffold | | Descriptor: | 1FD6 16055 V1V2 scaffold, 2-acetamido-2-deoxy-beta-D-glucopyranose, GN1_PA8 Fab Heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
6VJN
 
 | | Structure of NHP D11A.B5Fab in complex with 16055 V2b peptide | | Descriptor: | D11A.B5 Fab Heavy chain, D11A.B5 Fab Light chain, SODIUM ION, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
6WIT
 
 | | Crystal structure of NHP D15.SD7 Fab in complex with 16055 V1V2 1FD6 scaffold | | Descriptor: | 16055 V1V2 1FD6 Scaffold, 2-acetamido-2-deoxy-beta-D-glucopyranose, NHP GN1-SD7 Fab Heavy Chain, ... | | Authors: | Liban, T, Aljedani, S, Rodarte, J, Pancera, M. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
6WRP
 
 | | Crystal Structure of PI3-E12 Fab, An Antibody Against Human Parainfluenza Virus Type III | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Weidle, C, Pancera, M. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Protective antibodies against human parainfluenza virus type 3 infection.
Mabs, 13, 2021
|
|
7MU4
 
 | | Crystal Structure of HPV L1-directed D24.M01Fab | | Descriptor: | D24.M01 Fab Heavy Chain, D24.M01 Fab Light Chain, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterisation of Immune Responses to HPV Vaccination
To Be Published
|
|
6XOC
 
 | | Crystal structure of glVRC01 Fab in complex with anti-idiotypic iv4 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Aljedani, S.S, Weidle, C, Pancera, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of a VRC01-class germline targeting immunogen derived from anti-idiotypic antibodies.
Cell Rep, 35, 2021
|
|
6XLZ
 
 | | Structure of NHP D11A.F2 Fab in complex with 16055 V2b peptide | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Envelope glycoprotein gp160, NHP_D11A.F2_Fab_Heavy_chain, ... | | Authors: | Aljedani, S, Rodarte, J, Liban, T, Pancera, M. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
6XSN
 
 | |
7MX8
 
 | |
7MYT
 
 | |
6XE1
 
 | |
6C5V
 
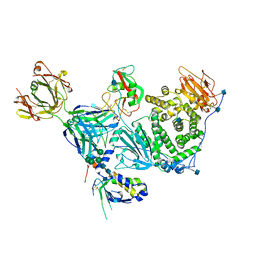 | | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab AMMO1 heavy chain, Antibody Fab AMMO1 light chain, ... | | Authors: | Snijder, J, Ortego, M.S, Weidle, C, Stuart, A.B, Gray, M.A, McElrath, M.J, Pancera, M, Veesler, D, McGuire, A.T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
2KM2
 
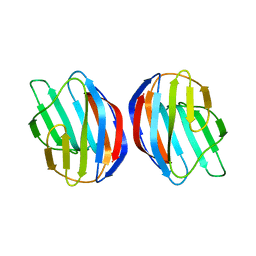 | | Galectin-1 dimer | | Descriptor: | Galectin-1 | | Authors: | Nesmelova, I.V, Ermakova, E, Daragan, V.A, Pang, M, Baum, L.G, Mayo, K.H. | | Deposit date: | 2009-07-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity.
J.Mol.Biol., 397, 2010
|
|
7KEO
 
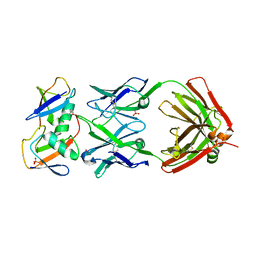 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7RQQ
 
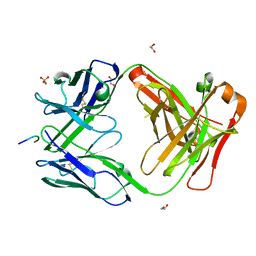 | |
7RQP
 
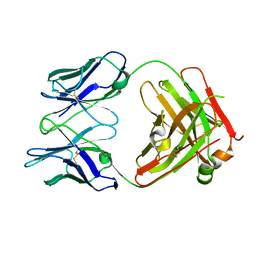 | | Structure of PfCSP NPNV binding antibody L9 | | Descriptor: | L9 Heavy Chain, L9 Kappa Chain | | Authors: | Hurlburt, N.K, Pancera, M. | | Deposit date: | 2021-08-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The light chain of the L9 antibody is critical for binding circumsporozoite protein minor repeats and preventing malaria.
Cell Rep, 38, 2022
|
|
7RQR
 
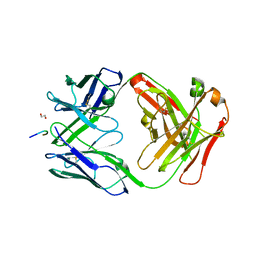 | |
3FRH
 
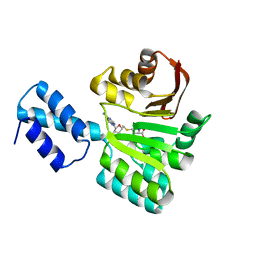 | | Structure of the 16S rRNA methylase RmtB, P21 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3FRI
 
 | | Structure of the 16S rRNA methylase RmtB, I222 | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schmitt, E, Galimand, M, Panvert, M, Dupechez, M, Courvalin, P, Mechulam, Y. | | Deposit date: | 2009-01-08 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural bases for 16 S rRNA methylation catalyzed by ArmA and RmtB methyltransferases
J.Mol.Biol., 388, 2009
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
5UTY
 
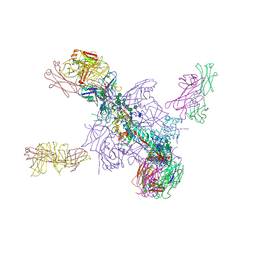 | | Crystal Structure of a Stabilized DS-SOSIP.mut4 BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, N300M, N302M, T320L in Complex with Human Antibodies PGT122 and 35O22 at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Fab heavy chain, ... | | Authors: | Xu, K, Chuang, G.-Y, Pancera, M, Kwong, P.D. | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.412 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
