6N3O
 
 | | Identification of novel, potent and selective GCN2 inhibitors as first-in-class anti-tumor agents | | Descriptor: | N-{3-[(2-aminopyrimidin-5-yl)ethynyl]-2,4-difluorophenyl}-5-chloro-2-methoxypyridine-3-sulfonamide, eIF-2-alpha kinase GCN2 | | Authors: | Hoffman, I.D, Fujimoto, J, Kurasawa, O, Takagi, T, Klein, M.G, Kefala, G, Ding, S.C, Cary, D.R, Mizojiri, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of Novel, Potent, and Orally Available GCN2 Inhibitors with Type I Half Binding Mode.
Acs Med.Chem.Lett., 10, 2019
|
|
8H2T
 
 | | Cryo-EM structure of IadD/E dioxygenase bound with IAA | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Aromatic-ring-hydroxylating dioxygenase beta subunit, FE (III) ION, ... | | Authors: | Yu, G, Li, Z, Zhang, H. | | Deposit date: | 2022-10-07 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural and biochemical characterization of the key components of an auxin degradation operon from the rhizosphere bacterium Variovorax.
Plos Biol., 21, 2023
|
|
6N77
 
 | | Structure of the human JAK1 kinase domain with compound 15 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N7D
 
 | | Structure of the human JAK1 kinase domain with compound 54 | | Descriptor: | GLYCEROL, N-[5-(6-methoxy-1H-indazol-5-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8HKG
 
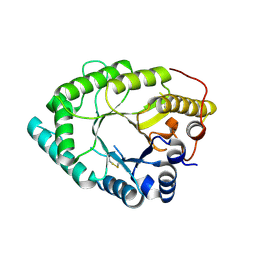 | |
8HX3
 
 | |
8I88
 
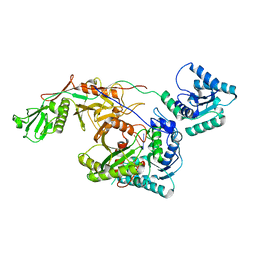 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I87
 
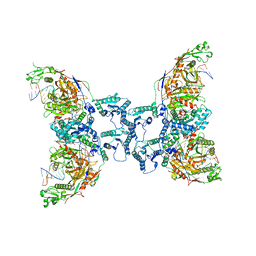 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Zhang, H, Deng, Z.Q, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
7WN8
 
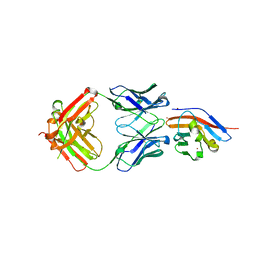 | | Crystal structure of antibody (BC31M5) binds to CD47 | | Descriptor: | BC31M5 Fab Heavy chain, BC31M5 Fab Light chain, Leukocyte surface antigen CD47, ... | | Authors: | Li, Y, Wang, W, Sui, J, Zhang, S. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A pH-dependent anti-CD47 antibody that selectively targets solid tumors and improves therapeutic efficacy and safety.
J Hematol Oncol, 16, 2023
|
|
3UBB
 
 | |
6KSF
 
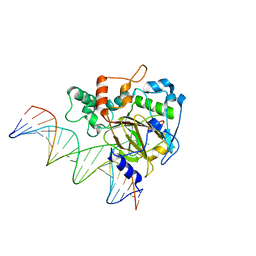 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | Authors: | Li, H, Zhang, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
3PWK
 
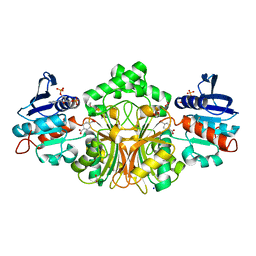 | | Crystal Structure of Aspartate beta-Semialdehide Dehydrogenase from Streptococcus pneumoniae with 2',5'-Adenosine diphosphate and D-2-aminoadipate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
3PZR
 
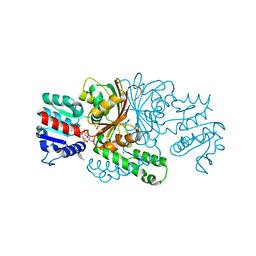 | | Crystals structure of aspartate beta-Semialdehyde dehydrogenase from Vibrio Cholerae with NADP and product of S-carbamoyl-L-cysteine | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CYSTEINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pavlovsky, A.G, Potente, N, Viola, R.E. | | Deposit date: | 2010-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
3Q11
 
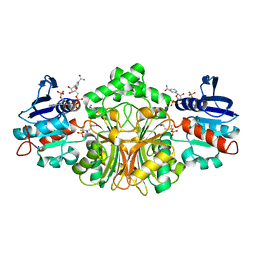 | | Crystals Structure of Aspartate beta-Semialdehyde Dehydrogenase from Streptococcus pneumoniae with NADP and aspartyl beta-difluorophosphonate | | Descriptor: | 1,2-ETHANEDIOL, 5,5-difluoro-4-oxo-5-phosphono-D-norvaline, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E, Faehnle, C.R. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
4E4K
 
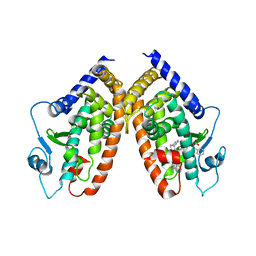 | | Crystal Structure of PPARgamma with the ligand JO21 | | Descriptor: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
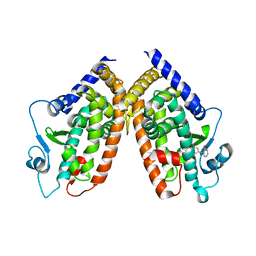 | | Crystal structure of PPARgamma with the ligand FS214 | | Descriptor: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
3WX8
 
 | |
4E1N
 
 | |
4E1M
 
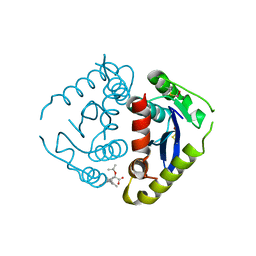 | |
4E7Z
 
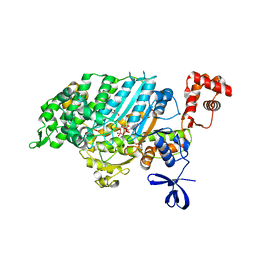 | | Myosin VI (MD) pre-powerstroke state, P21 crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Isabet, T, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin VI.
Mol.Cell, 48, 2012
|
|
6IP0
 
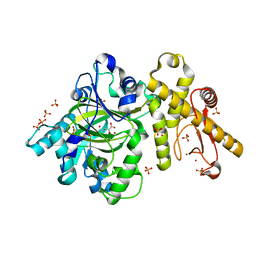 | |
6IP4
 
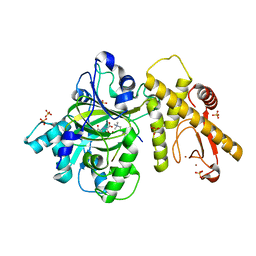 | |
4E7S
 
 | | Myosin VI D23R I24R R569E (MD) pre-powerstroke state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Isabet, T, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2012-03-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Processive Steps in the Reverse Direction Require Uncoupling of the Lead Head Lever Arm of Myosin VI.
Mol.Cell, 48, 2012
|
|
8IPJ
 
 | | Crystal structure of the Legionella effector protein MavL with ADPR-Ub | | Descriptor: | MavL, Ubiquitin, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Legionella maintains host cell ubiquitin homeostasis by effectors with unique catalytic mechanisms.
Nat Commun, 15, 2024
|
|
8IPW
 
 | | The sturecture of Legionella effector protein MavL with ADPR | | Descriptor: | MavL, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL[HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H. | | Deposit date: | 2023-03-15 | | Release date: | 2024-03-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Legionella maintains host cell ubiquitin homeostasis by effectors with unique catalytic mechanisms.
Nat Commun, 15, 2024
|
|
