6LWR
 
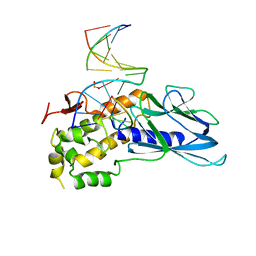 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing a cleaved C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*(PDA))-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
5N1Y
 
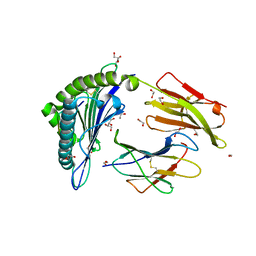 | | HLA-A02 carrying MVWGPDPLYV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2017-02-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J. Clin. Invest., 126, 2016
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
6ALC
 
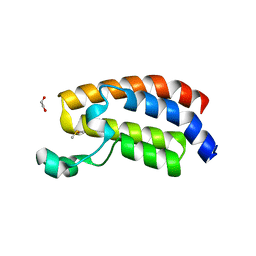 | | CREBBP bromodomain in complex with Cpd 4 (1-(1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]ethan-1-one, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Design and synthesis of a biaryl series as inhibitors for the bromodomains of CBP/P300.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6PAV
 
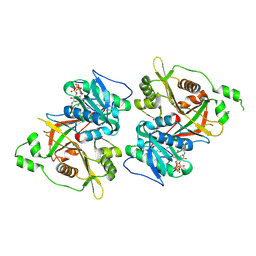 | |
4QXW
 
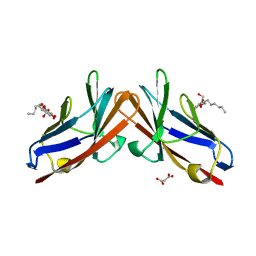 | | Crystal structure of the human CEACAM1 membrane distal amino terminal (N)-domain | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Huang, Y.H, Gandhi, A.K, Russell, A, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2014-07-22 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
6PAU
 
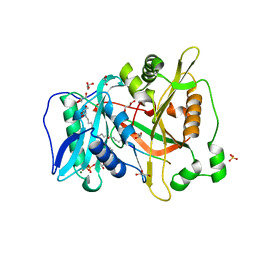 | |
8H7B
 
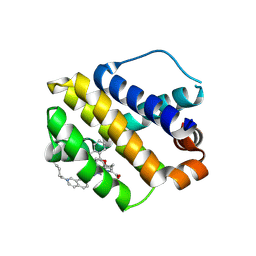 | | The crystal structure of human mcl1 kinase domain in complex with MCL1-M-EBA | | Descriptor: | 7-[3-(isoquinolin-7-yloxymethyl)-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46408451 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
8H7F
 
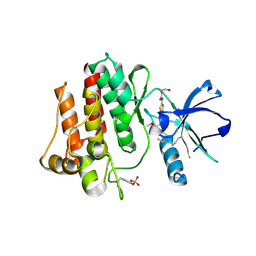 | | The crystal structure of human abl1 kinase domain in complex with abl1-B-EBA | | Descriptor: | 1-[6-(6-methoxyisoquinolin-7-yl)-1,3-benzothiazol-2-yl]-3-(2-oxidanylideneethyl)urea, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45013428 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
8H7H
 
 | | The crystal structure of human abl1 kinase domain in complex with abl1-A-EBA | | Descriptor: | 5-[3-(6-methoxyisoquinolin-7-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N-methyl-N-prop-2-ynyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C.J, Zhang, Z.M. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27789712 Å) | | Cite: | 2-Ethynylbenzaldehyde-Based, Lysine-Targeting Irreversible Covalent Inhibitors for Protein Kinases and Nonkinases.
J.Am.Chem.Soc., 2023
|
|
8JF4
 
 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | Descriptor: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | Authors: | Zhu, C.J. | | Deposit date: | 2023-05-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89288354 Å) | | Cite: | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5C0G
 
 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5C0I
 
 | | HAL-A02 carrying RQFGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5C0E
 
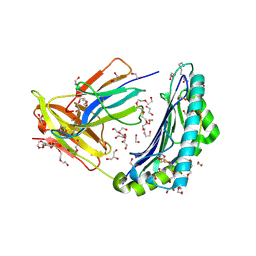 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
5DZL
 
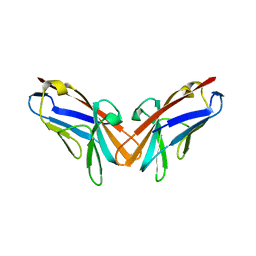 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
5UK5
 
 | | Complex of Notch1(EGF8-12) bound to Jagged1(N-EGF3) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Garcia, K.C, Luca, V.C. | | Deposit date: | 2017-01-19 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity.
Science, 355, 2017
|
|
4M5F
 
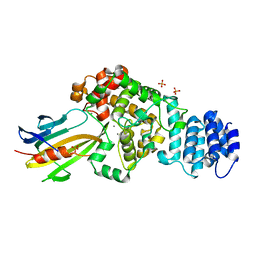 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N88
 
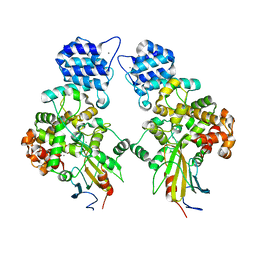 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
6K5K
 
 | |
6K8P
 
 | |
4N80
 
 | | Crystal structure of Tse3-Tsi3 complex | | Descriptor: | CALCIUM ION, Uncharacterized protein, ZINC ION | | Authors: | Shang, G.J. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
6K5G
 
 | |
8JWY
 
 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
