7LWE
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-7629 | | Descriptor: | ACETATE ION, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWG
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12694 | | Descriptor: | 5-{(5S)-1-[2-({3-chloro-6-[(2S)-2,4-dimethylpiperazin-1-yl]-2-fluoropyridin-4-yl}amino)-2-oxoethyl]-4-oxo-4,6,7,8-tetrahydro-1H-dipyrrolo[1,2-a:2',3'-d]pyrimidin-3-yl}-3,4-difluoro-2-hydroxybenzamide, B-cell lymphoma 6 protein, GLYCEROL, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWF
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-9320 | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, N-(3-chloropyridin-4-yl)-2-[5-(3-cyano-4-hydroxyphenyl)-3-methyl-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LZQ
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-4425 | | Descriptor: | B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, N-(3-chloropyridin-4-yl)-2-(5-methyl-4-oxo-4,5-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
4OUC
 
 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
8CAO
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with CA24 | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-(4-cyclohexylphenyl)quinolin-4-yl]carbonyl-1,3,8-triazaspiro[4.5]decane-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CB0
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CAP
 
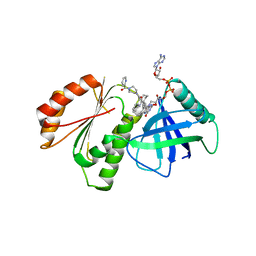 | |
8CAL
 
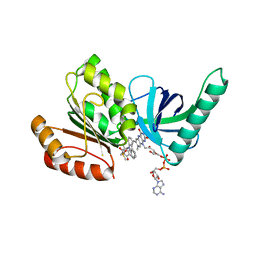 | |
8CAK
 
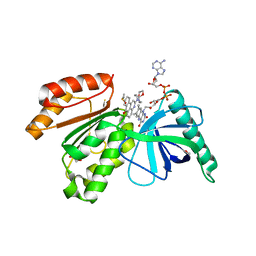 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, DIMETHYL SULFOXIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
6TUY
 
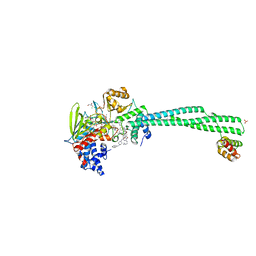 | | Human LSD1/CoREST bound to the quinazoline inhibitor MC4106 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mattevi, A, Marrocco, B. | | Deposit date: | 2020-01-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel non-covalent LSD1 inhibitors endowed with anticancer effects in leukemia and solid tumor cellular models.
Eur.J.Med.Chem., 237, 2022
|
|
3U78
 
 | | E67-2 selectively inhibits KIAA1718, a human histone H3 lysine 9 Jumonji demethylase | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, ... | | Authors: | Upadhyay, A.K, Cheng, X. | | Deposit date: | 2011-10-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | An Analog of BIX-01294 Selectively Inhibits a Family of Histone H3 Lysine 9 Jumonji Demethylases.
J.Mol.Biol., 416, 2012
|
|
6GOU
 
 | | Development of Alkyl Glycerone Phosphate Synthase Inhibitors: Complex with Inhibitor 2I | | Descriptor: | (3~{S})-3-[2,6-bis(fluoranyl)phenyl]-~{N}-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)methyl]butanamide, Alkyldihydroxyacetonephosphate synthase, peroxisomal, ... | | Authors: | Mattevi, A, Piano, V. | | Deposit date: | 2018-06-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of alkyl glycerone phosphate synthase inhibitors: Structure-activity relationship and effects on ether lipids and epithelial-mesenchymal transition in cancer cells.
Eur J Med Chem, 163, 2018
|
|
5MAR
 
 | | Structure of human SIRT2 in complex with 1,2,4-Oxadiazole inhibitor and ADP ribose. | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]propan-1-ol, ACETATE ION, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure-Activity Relationship, X-ray Crystal Structure, and Anticancer Activity.
J. Med. Chem., 60, 2017
|
|
6XVW
 
 | |
5MFP
 
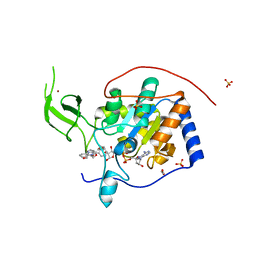 | | Human Sirt6 in complex with activator UBCS58 | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridin-3-ylpyrrolo[1,2-a]quinoxaline, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MF6
 
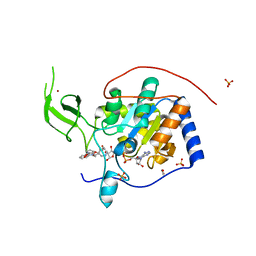 | | Human Sirt6 in complex with activator UBCS039 | | Descriptor: | (4~{R})-4-pyridin-3-yl-4,5-dihydropyrrolo[1,2-a]quinoxaline, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MFZ
 
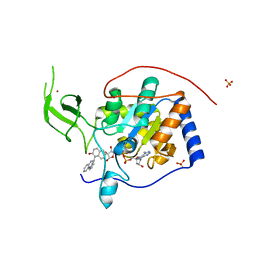 | | Human Sirt6 in complex with small molecule UBCS40 | | Descriptor: | 1,2-ETHANEDIOL, 1-pyrrolo[1,2-a]quinoxalin-4-ylnaphthalen-2-ol, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-19 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MGN
 
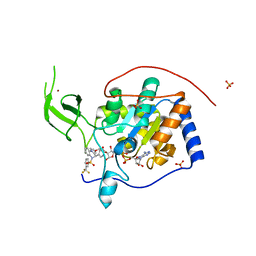 | | Human Sirt6 in complex with activator UBCS38 | | Descriptor: | (4~{R})-4-pyridin-3-yl-5-[3-(trifluoromethyl)phenyl]sulfonyl-4~{H}-pyrrolo[1,2-a]quinoxaline, 1,2-ETHANEDIOL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Steegborn, C, You, W, Kambach, C. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8FS1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8CBH
 
 | | SHP2 in complex with a novel allosteric inhibitor | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [(1~{S},6~{R},7~{S})-3-[3-[2,3-bis(chloranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyrazin-6-yl]-7-(4-methyl-1,3-thiazol-2-yl)-3-azabicyclo[4.1.0]heptan-7-yl]methanamine | | Authors: | di Fabio, R, Petrocchi, A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
