1CBO
 
 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES HIS447ASN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-02-26 | | Release date: | 1999-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6Y4W
 
 | | Crystal structure of p38 in complex with SR69 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(cyclohexylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6W75
 
 | | 1.95 Angstrom Resolution Crystal Structure of NSP10 - NSP16 Complex from SARS-CoV-2 | | Descriptor: | 2'-O-methyltransferase, FORMIC ACID, Non-structural protein 10, ... | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Wiersum, G, Godzik, A, Jaroszewski, L, Stogios, P.J, Skarina, T, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
7KSB
 
 | | Crystal structure on Act c 10.0101 | | Descriptor: | Non-specific lipid-transfer protein 1, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Giangrieco, I, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
2VME
 
 | | Structure of the wild-type discoidin II from Dictyostelium discoideum | | Descriptor: | CALCIUM ION, CHLORIDE ION, DISCOIDIN-2, ... | | Authors: | Aragao, K.S, Satre, M, Imberty, A, Varrot, A. | | Deposit date: | 2008-01-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure Determination of Discoidin II from Dictyostelium Discoideum and Carbohydrate Binding Properties of the Lectin Domain.
Proteins, 73, 2008
|
|
6NSO
 
 | | An Unexpected Intermediate in the Reaction Catalyzed by Quinolinate Synthase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, IRON/SULFUR CLUSTER, Quinolinate synthase A | | Authors: | Esakova, O.A, Grove, T.L, Silakov, A, Yennawar, N.H, Booker, S.J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Unexpected Species Determined by X-ray Crystallography that May Represent an Intermediate in the Reaction Catalyzed by Quinolinate Synthase.
J.Am.Chem.Soc., 141, 2019
|
|
6NT3
 
 | | Cryo-EM structure of a human-cockroach hybrid Nav channel. | | Descriptor: | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clairfeuille, T, Rohou, A, Payandeh, J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-02-20 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a human-cockroach hybrid Nav channel in the presence and absence of the alpha-scorpion toxin AaH2.
Science, 2019
|
|
3M8S
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in a closed ternary complex with trapped 4'-methylated dTTP | | Descriptor: | 4'-methylthymidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederichs, K, Marx, A, Betz, K. | | Deposit date: | 2010-03-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of DNA polymerases caught processing size-augmented nucleotide probes.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6Y6F
 
 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
1EL3
 
 | | HUMAN ALDOSE REDUCTASE COMPLEXED WITH IDD384 INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,6-DIMETHYL-4-(2-O-TOLYL-ACETYLAMINO)-BENZENESULFONYL]-GLYCINE | | Authors: | Podjarny, A. | | Deposit date: | 2000-03-13 | | Release date: | 2000-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of human aldose reductase bound to the inhibitor IDD384.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
8OKV
 
 | | lipoprotein BT2095 from Bacteroides thetaiotamicron bound to cyanocobalamin CnCbl | | Descriptor: | CYANOCOBALAMIN, Putative surface layer protein | | Authors: | Javier Abellon-Ruiz, J, van den Berg, B, Jana, K, Kleinekathofer, U, Silale, A, Frey, A.M, Basle, A, Trost, M. | | Deposit date: | 2023-03-29 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
1CBK
 
 | | 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | 7,8-DIHYDRO-7,7-DIMETHYL-6-HYDROXYPTERIN, PROTEIN (7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE), SULFATE ION | | Authors: | Hennig, M, D'Arcy, A, Dale, G, Oefner, C. | | Deposit date: | 1999-02-26 | | Release date: | 2000-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The structure and function of the 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Haemophilus influenzae.
J.Mol.Biol., 287, 1999
|
|
6NXJ
 
 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6TIG
 
 | | Structure of the N terminal domain of Bc2L-C lectin (1-131) in complex with Globo H (H-type 3) antigen | | Descriptor: | Lectin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Varrot, A, Bermeo, R. | | Deposit date: | 2019-11-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BC2L-C N-Terminal Lectin Domain Complexed with Histo Blood Group Oligosaccharides Provides New Structural Information.
Molecules, 25, 2020
|
|
6NB8
 
 | | Crystal structure of anti- SARS-CoV human neutralizing S230 antibody Fab fragment | | Descriptor: | S230 antigen-binding (Fab) fragment, heavy chain, light chain | | Authors: | Walls, A.J, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, J, Quispe, J, Cameroni, E, Gopal, R, Dai, M, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
6NBT
 
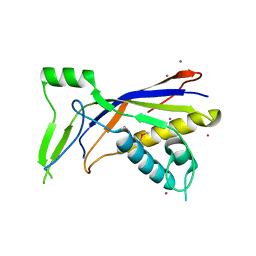 | |
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
1CDI
 
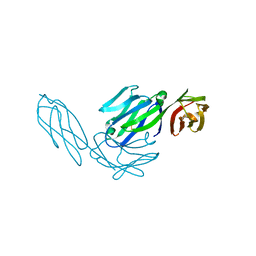 | |
5X7S
 
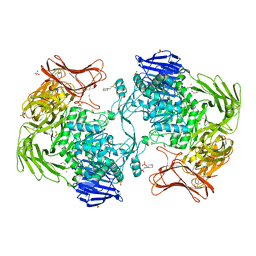 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2UV4
 
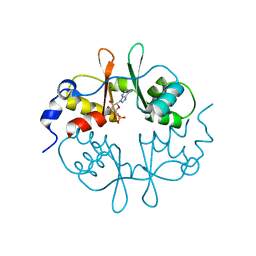 | | Crystal Structure of a CBS domain pair from the regulatory gamma1 subunit of human AMPK in complex with AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ADENOSINE MONOPHOSPHATE | | Authors: | Day, P, Sharff, A, PArra, L, Cleasby, A, Williams, M, Horer, S, Nar, H, Redemann, N, Tickle, I, Yon, J. | | Deposit date: | 2007-03-09 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure of a Cbs-Domain Pair from the Regulatory Gamma1 Subunit of Human Ampk in Complex with AMP and Zmp.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
7KG3
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | Authors: | Arvai, A, Brosey, C.A, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7F3O
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-653 | | Descriptor: | 7-(4-cyclohexyloxyphenyl)-9-methyl-4$l^{6}-thia-1$l^{4},5,8-triazabicyclo[4.4.0]deca-1(10),6,8-triene 4,4-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2021-06-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strictly regulated agonist-dependent activation of AMPA-R is the key characteristic of TAK-653 for robust synaptic responses and cognitive improvement.
Sci Rep, 11, 2021
|
|
