1RZ4
 
 | | Crystal Structure of Human eIF3k | | Descriptor: | Eukaryotic translation initiation factor 3 subunit 11, SULFATE ION | | Authors: | Wei, Z, Zhang, P, Zhou, Z, Gong, W. | | Deposit date: | 2003-12-23 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human eIF3k, the first structure of eIF3 subunits
J.Biol.Chem., 279, 2004
|
|
1XRF
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | | Descriptor: | Dihydroorotase, SULFATE ION, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
5E8H
 
 | | Crystal structure of geranylfarnesyl pyrophosphate synthases 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 3, chloroplastic | | Authors: | Wang, C, Chen, Q, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.028 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8L
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | Descriptor: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
1XRT
 
 | | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex Aeolicus at 1.7 A Resolution | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Martin, P.D, Purcarea, C, Zhang, P, Vaishnav, A, Sadecki, S, Guy-Evans, H.I, Evans, D.R, Edwards, B.F. | | Deposit date: | 2004-10-15 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | The crystal structure of a novel, latent dihydroorotase from Aquifex aeolicus at 1.7A resolution
J.Mol.Biol., 348, 2005
|
|
3IWP
 
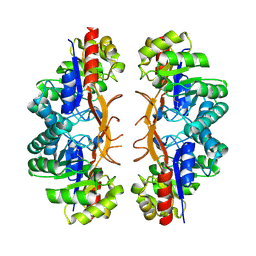 | |
3J4F
 
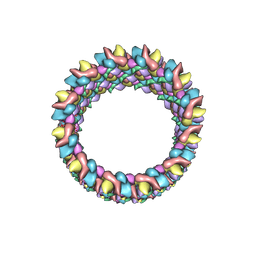 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J3Y
 
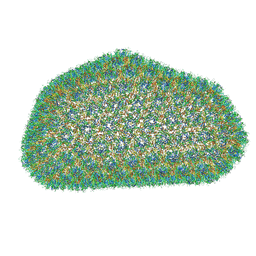 | |
3JA6
 
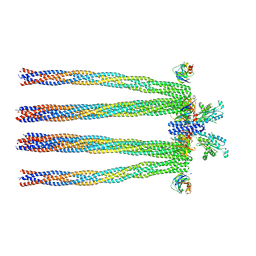 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
3J3Q
 
 | |
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6BYS
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PRKAc alpha | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.75 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
6BYR
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
5XN5
 
 | |
5X3X
 
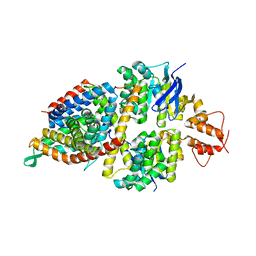 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
7XTQ
 
 | | Cryo-EM structure of the R399-bound GPBAR-Gs complex | | Descriptor: | G-protein coupled bile acid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, L, Yang, F, Wu, X, Mao, C, Sun, J, Yu, X, Zhang, Y, Zhang, P. | | Deposit date: | 2022-05-17 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis and molecular mechanism of biased GPBAR signaling in regulating NSCLC cell growth via YAP activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5X41
 
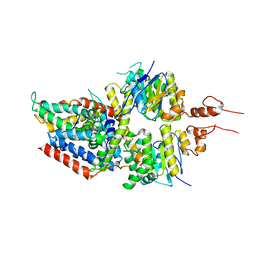 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
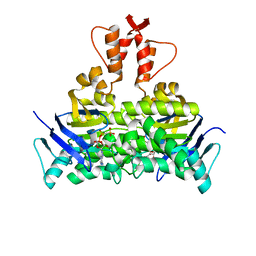 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | Descriptor: | Os02g0668100 protein, Os07g0580900 protein | | Authors: | Wang, C, Zhou, F, Lu, S, Zhang, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7X0X
 
 | |
7X0Y
 
 | |
3P4F
 
 | | Structural and biochemical insights into MLL1 core complex assembly and regulation. | | Descriptor: | Histone-lysine N-methyltransferase MLL, Retinoblastoma-binding protein 5, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Groulx, A, Tremblay, V, Brunzelle, J.B, Couture, J.-F. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical insights into MLL1 core complex assembly.
Structure, 19, 2011
|
|
3PSL
 
 | | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3 based peptide mimetic | | Descriptor: | N-alpha acetylated form of histone H3, WD repeat-containing protein 5 | | Authors: | Avdic, V, Zhang, P, Lanouette, S, Voronova, A, Skerjanc, I, Couture, J.-F. | | Deposit date: | 2010-12-01 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fine-tuning the stimulation of MLL1 methyltransferase activity by a histone H3-based peptide mimetic.
Faseb J., 25, 2011
|
|
3S32
 
 | | Crystal structure of Ash2L N-terminal domain | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Sarvan, S, Avdic, V, Tremblay, V, Chaturvedi, C.-P, Zhang, P, Lanouette, S, Blais, A, Brunzelle, J.S, Brand, M, Couture, J.-F. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-08 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
