6ZOM
 
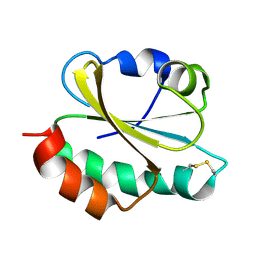 | | Oxidized thioredoxin 1 from the anaerobic bacteria Desulfovibrio vulgaris Hildenborough | | Descriptor: | Thioredoxin | | Authors: | Garcin, E, Bornet, O, Nouailler, M, Pieulle, L, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system.
Extremophiles, 25, 2021
|
|
6N4C
 
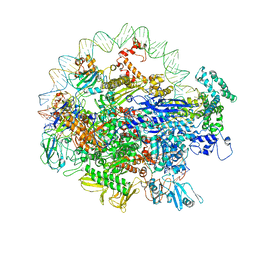 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
7OLY
 
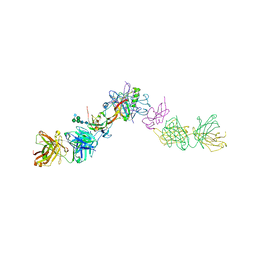 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
5BN2
 
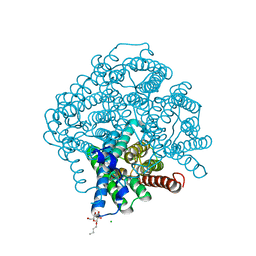 | | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A | | Descriptor: | AQY1 protein, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fischer, G, Kosinska Eriksson, U, Hedfalk, K, Neutze, R. | | Deposit date: | 2015-05-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A
To Be Published
|
|
3TT0
 
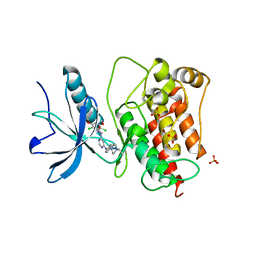 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
7TDZ
 
 | |
3ZOJ
 
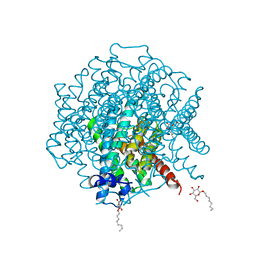 | | High-resolution structure of Pichia Pastoris aquaporin Aqy1 at 0.88 A | | Descriptor: | AQUAPORIN, CHLORIDE ION, octyl beta-D-glucopyranoside | | Authors: | Kosinska-Eriksson, U, Fischer, G, Friemann, R, Enkavi, G, Tajkhorshid, E, Neutze, R. | | Deposit date: | 2013-02-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Subangstrom Resolution X-Ray Structure Details Aquaporin-Water Interactions
Science, 340, 2013
|
|
8P0M
 
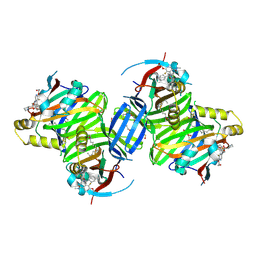 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 2024
|
|
3NA4
 
 | | D53P beta-2 microglobulin mutant | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Beta-2-microglobulin | | Authors: | Azinas, S, Ricagno, S, Bolognesi, M. | | Deposit date: | 2010-06-01 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-strand perturbation and amyloid propensity in beta-2 microglobulin
Febs J., 278, 2011
|
|
8E3G
 
 | | BMP2/GDF5 heterodimer | | Descriptor: | Bone morphogenetic protein 2, GLYCEROL, Growth/differentiation factor 5 | | Authors: | Gipson, G.R, Nolan, K.T, Thompson, T.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Formation and characterization of BMP2/GDF5 and BMP4/GDF5 heterodimers.
Bmc Biol., 21, 2023
|
|
1B6I
 
 | | T4 LYSOZYME MUTANT WITH CYS 54 REPLACED BY THR, CYS 97 REPLACED BY ALA, THR 21 REPLACED BY CYS AND LYS 124 REPLACED BY CYS (C54T,C97A,T21C,K124C) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, PROTEIN (LYSOZYME) | | Authors: | Vetter, I.R, Baase, W.A, Snow, S, Matthews, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3UM8
 
 | | Wild-type Plasmodium falciparum DHFR-TS complexed with cycloguanil and NADPH | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3UM6
 
 | | Double mutant (A16V+S108T) Plasmodium falciparum DHFR-TS (T9/94) complexed with cycloguanil, NADPH and dUMP | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
3SEK
 
 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
4KUP
 
 | | Endothiapepsin in complex with 20mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3UM5
 
 | | Double mutant (A16V+S108T) Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-T9/94) complexed with pyrimethamine, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Kamchonwongpaisan, S, Chitnumsub, P, Yuthavong, Y. | | Deposit date: | 2011-11-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Spatial Limitation around Residues 16 and 108 of Plasmodium falciparum Dihydrofolate Reductase Explains Resistance to Cycloguanil.
Antimicrob.Agents Chemother., 56, 2012
|
|
4L3C
 
 | | Structure of HLA-A2 in complex with D76N b2m mutant and NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
4LBT
 
 | | Endothiapepsin in complex with 100mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Winquist, J, Klebe, G. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4LHH
 
 | | Endothiapepsin in complex with 2mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2013-07-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4L29
 
 | | Structure of wtMHC class I with NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-04 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
5EL2
 
 | |
5HK5
 
 | |
5HCT
 
 | | Endothiapepsin in complex with biacylhydrazone | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-N'-{3-[(E)-{2-[(2S)-2-amino-3-(1H-indol-3-yl)propanoyl]hydrazinylidene}methyl]benzylidene}-3-(1H-indol-2-yl)propanehydrazide (non-preferred name), ACETATE ION, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2016-01-04 | | Release date: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment Linking and Optimization of Inhibitors of the Aspartic Protease Endothiapepsin: Fragment-Based Drug Design Facilitated by Dynamic Combinatorial Chemistry.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
