8JQB
 
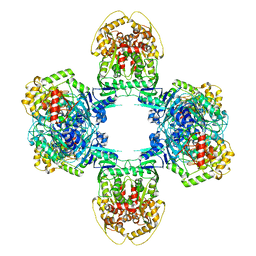 | | Structure of Gabija GajA-GajB 4:4 Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8JQC
 
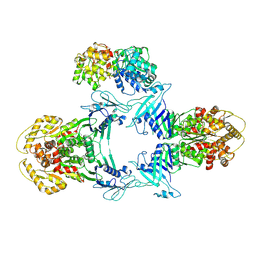 | | Novel Anti-phage System | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Li, J, Wang, Z, Wang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
6JQH
 
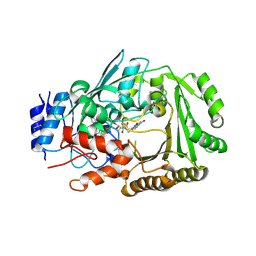 | | Crystal structure of MaDA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MaDA | | Authors: | Du, X.X, Lei, X.G. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis.
Nat.Chem., 12, 2020
|
|
6LQE
 
 | |
6LQF
 
 | |
3IJJ
 
 | | Ternary Complex of Macrophage Migration Inhibitory Factor (MIF) Bound Both to 4-hydroxyphenylpyruvate and to the Allosteric Inhibitor AV1013 (R-stereoisomer) | | Descriptor: | (2E)-2-hydroxy-3-(4-hydroxyphenyl)prop-2-enoic acid, (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ... | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IJG
 
 | | Macrophage Migration Inhibitory Factor (MIF) Bound to the (R)-Stereoisomer of AV1013 | | Descriptor: | (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3F5P
 
 | | Complex Structure of Insulin-like Growth Factor Receptor and 3-Cyanoquinoline Inhibitor | | Descriptor: | 4-[[3-chloro-4-(1-methylimidazol-2-yl)sulfanyl-phenyl]amino]-7-[3-(2-hydroxyethyl-methyl-amino)propoxy]-6-methoxy-quinoline-3-carbonitrile, Insulin-like growth factor 1 receptor | | Authors: | Xu, W, Miller, L.M, Mayer, S.C, Berger, D.M, Boschelli, D.H, Boschelli, F. | | Deposit date: | 2008-11-04 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lead identification to generate 3-cyanoquinoline inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GRW
 
 | | FGFR3 in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Wiesmann, C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice.
J.Clin.Invest., 119, 2009
|
|
3RF4
 
 | | Ancylostoma ceylanicum mif in complex with furosemide | | Descriptor: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, ACETATE ION, IMIDAZOLE, ... | | Authors: | Cho, Y, Lolis, E. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Drug Repositioning and Pharmacophore Identification in the Discovery of Hookworm MIF Inhibitors.
Chem.Biol., 18, 2011
|
|
3RF5
 
 | | Ancylostoma ceylanicum mif in complex with n-(2,3,4,5,6-pentafluoro-benzyl)-4-sulfamoyl-benzamide | | Descriptor: | 2-[(furan-2-ylmethyl)amino]benzoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Cho, Y, Lolis, E. | | Deposit date: | 2011-04-05 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug Repositioning and Pharmacophore Identification in the Discovery of Hookworm MIF Inhibitors.
Chem.Biol., 18, 2011
|
|
5ZM7
 
 | | Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
5ZM6
 
 | | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 | | Descriptor: | ACETATE ION, Oxysterol-binding protein-related protein 1, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Dong, J, Wang, J, Luo, Z, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
5ZM5
 
 | | Crystal structure of human ORP1-ORD in complex with cholesterol at 2.6 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
8SKL
 
 | | PTP1B in complex with 182 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-fluoro-3-hydroxy-7-(3-hydroxy-3-methylbutoxy)naphthalen-2-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, ... | | Authors: | Babon, J.J, Chen, H, Tiganis, T. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A small molecule inhibitor of PTP1B and PTPN2 enhances T cell anti-tumor immunity.
Nat Commun, 14, 2023
|
|
8P1X
 
 | | TarM(Se)_G117R-UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2023-05-13 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
8P20
 
 | | TarM(Se)_G117R-UDP-4RboP-glucose | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.848 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
5U77
 
 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
4N79
 
 | | Structure of Cathepsin K-dermatan sulfate complex | | Descriptor: | Cathepsin K, alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of collagen fiber degradation by cathepsin K.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5U78
 
 | | Crystal structure of ORP8 PH domain in P1211 space group | | Descriptor: | Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-12 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
4N8W
 
 | | cathepsin K - chondroitin sulfate complex | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Cathepsin K | | Authors: | Aguda, A.H, Nguyen, N.T, Bromme, D, Brayer, G.D. | | Deposit date: | 2013-10-18 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of collagen fiber degradation by cathepsin K.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7QD7
 
 | | TarM(Se)_G117R | | Descriptor: | CHLORIDE ION, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-11-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QH9
 
 | | TarM(Se)_G117R-4RboP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TarM(Se)_G117R-4RboP, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-10 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
7QNT
 
 | | TarM(Se) native | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2021-12-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Invasive Staphylococcus epidermidis uses a unique processive wall teichoic acid glycosyltransferase to evade immune recognition.
Sci Adv, 9, 2023
|
|
4JAN
 
 | | crystal structure of broadly neutralizing antibody CH103 in complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
