3ER9
 
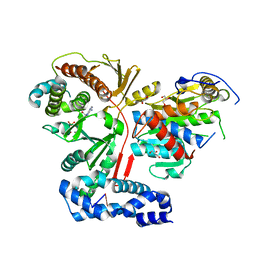 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with UU and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-R(UP*U)-3', CALCIUM ION, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3ER8
 
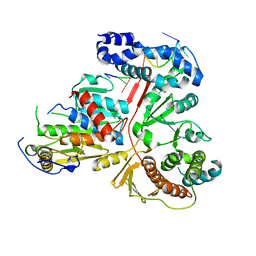 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
4WLG
 
 | |
4WMB
 
 | |
4WMK
 
 | |
4WMI
 
 | |
4WM0
 
 | |
4WN2
 
 | |
4WMA
 
 | |
5TRR
 
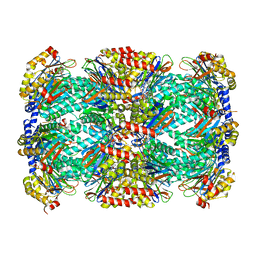 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
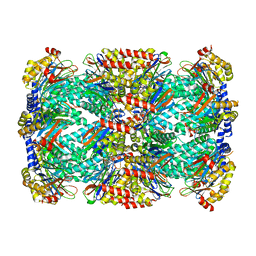 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TS0
 
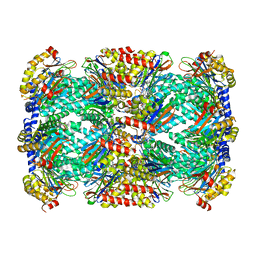 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRS
 
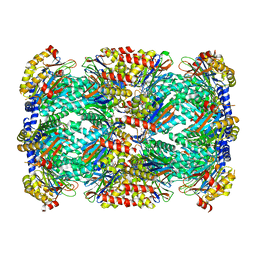 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRG
 
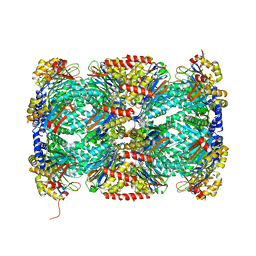 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
1Q2O
 
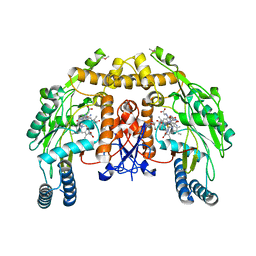 | | Bovine endothelial nitric oxide synthase N368D mutant heme domain dimer with L-N(omega)-nitroarginine-2,4-L-diaminobutyramide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-07-25 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6XN5
 
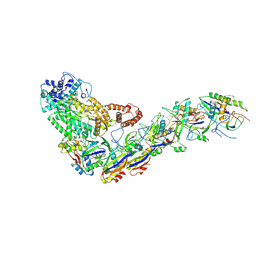 | | Structure of the Lactococcus lactis Csm Apo- CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm3, CRISPR-associated protein Csm4, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN3
 
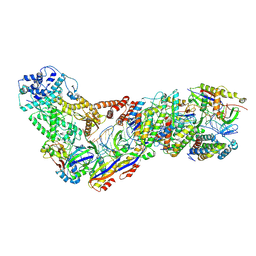 | | Structure of the Lactococcus lactis Csm CTR_4:3 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN4
 
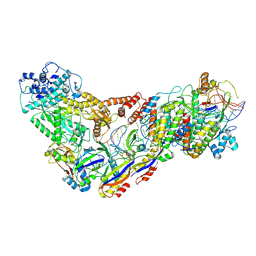 | | Structure of the Lactococcus lactis Csm CTR_3:2 CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
6XN7
 
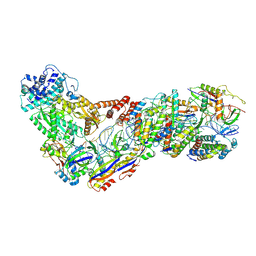 | | Structure of the Lactococcus lactis Csm NTR CRISPR-Cas Complex | | Descriptor: | CRISPR-associated protein Cas10, CRISPR-associated protein Csm2, CRISPR-associated protein Csm3, ... | | Authors: | Rai, J, Sridhara, S, Li, H. | | Deposit date: | 2020-07-02 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural and biochemical characterization of in vivo assembled Lactococcus lactis CRISPR-Csm complex.
Commun Biol, 5, 2022
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
5U8T
 
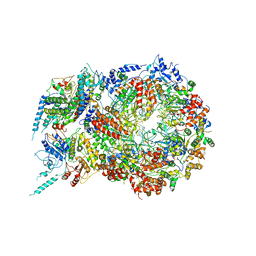 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5THO
 
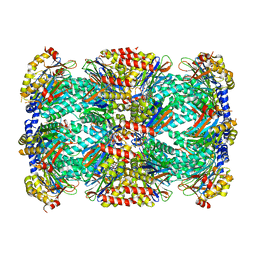 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome in complex with N,C-capped Dipeptide Inhibitor PKS2205 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2016-09-30 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
2VQI
 
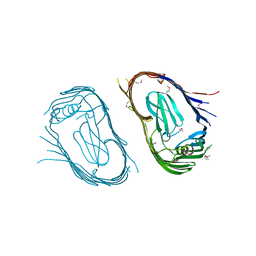 | | Structure of the P pilus usher (PapC) translocation pore | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE USHER PROTEIN PAPC | | Authors: | Remaut, H, Tang, C, Henderson, N.S, Pinkner, J.S, Wang, T, Hultgren, S.J, Thanassi, D.G, Li, H, Waksman, G. | | Deposit date: | 2008-03-16 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Fiber formation across the bacterial outer membrane by the chaperone/usher pathway.
Cell, 133, 2008
|
|
1I83
 
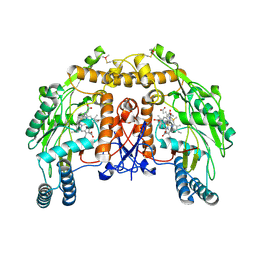 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH N1,N14-BIS((S-METHYL)ISOTHIOUREIDO)TETRADECANE (H4B FREE) | | Descriptor: | ACETATE ION, GLYCEROL, N1,N14-BIS((S-METHYL)ISOTHIOUREIDO)TETRADECANE, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Babu, B.R, Griffith, O.W, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase.
J.Biol.Chem., 276, 2001
|
|
6OWU
 
 | |
