8TUH
 
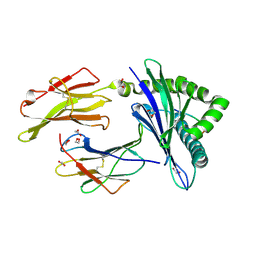 | | HLA B7:02 with RPIIRPATL | | Descriptor: | 1,2-ETHANEDIOL, ARG-PRO-ILE-ILE-ARG-PRO-ALA-THR-LEU, Beta-2-microglobulin, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.200106 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8TUB
 
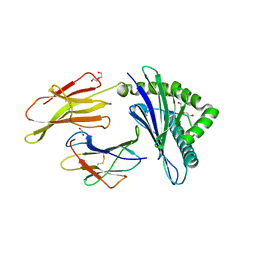 | | HLA B7:02 with HPNGYKSLSTL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8TWQ
 
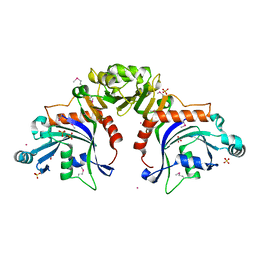 | | Structure of bacteriophage lambda RexA protein | | Descriptor: | CADMIUM ION, Protein rexA, SULFATE ION | | Authors: | Adams, M.C, Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of bacteriophage lambda RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins.
Nucleic Acids Res., 52, 2024
|
|
8TTQ
 
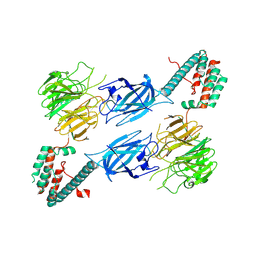 | |
8UMO
 
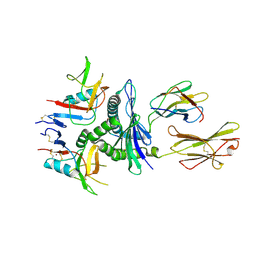 | | Murine CD94-NKG2A receptor in complex with Qa-1b presenting AMAPRTLLL | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2023-10-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the murine CD94-NKG2A receptor in complex with Qa-1 b presenting an MHC-I leader peptide.
Febs J., 291, 2024
|
|
2FL2
 
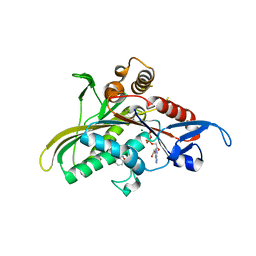 | | crystal structure of KSP in complex with inhibitor 19 | | Descriptor: | (1S)-1-CYCLOPROPYL-2-[(2S)-4-(2,5-DIFLUOROPHENYL)-2-PHENYL-2,5-DIHYDRO-1H-PYRROL-1-YL]-2-OXOETHANAMINE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FL6
 
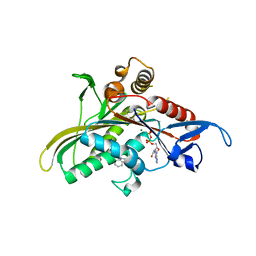 | | crystal structure of KSP in complex with inhibitor 6 | | Descriptor: | (2S)-4-(2,5-DIFLUOROPHENYL)-N,N-DIMETHYL-2-PHENYL-2,5-DIHYDRO-1H-PYRROLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 2: the design, synthesis, and characterization of 2,4-diaryl-2,5-dihydropyrrole inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
2G1Q
 
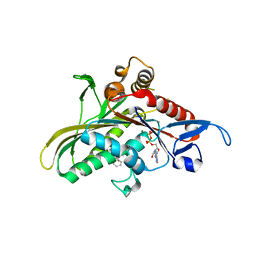 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7VO8
 
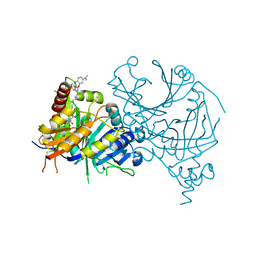 | | Structure of AtHPPD complexed with LSY-1 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-[6-[[9-(dimethylamino)-12H-benzo[a]phenoxazin-5-yl]amino]hexyl]-4-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[oxidanyl(oxidanylidene)-$l^4-azanyl]benzamide | | Authors: | Yang, G.-F, Dong, J. | | Deposit date: | 2021-10-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In vivo diagnostics of abiotic plant stress responses via in situ real-time fluorescence imaging
Plant Physiol., 190, 2022
|
|
7VLG
 
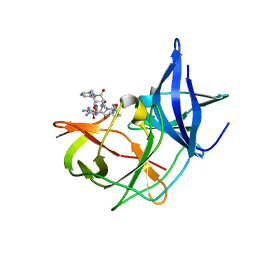 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2201 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-5-propan-2-yl-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLH
 
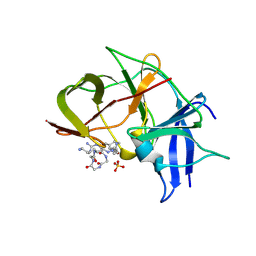 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2219 | | Descriptor: | 1-[(3~{S},6~{R},18~{R})-3,6-bis(4-azanylbutyl)-2,5,8,11,14,17-hexakis(oxidanylidene)-1,4,7,10,13,16-hexazacyclodocos-18-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7UDW
 
 | | Designed pentameric proton channel QQLL | | Descriptor: | De novo designed pentameric proton channel QQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDV
 
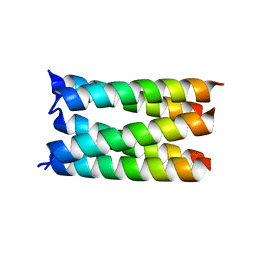 | |
7UDZ
 
 | | Designed pentameric proton channel LQLL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDY
 
 | | Designed pentameric channel QLLL | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDX
 
 | | Designed pentameric proton channel QLQL | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel QLQL | | Authors: | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | Deposit date: | 2022-03-20 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UPI
 
 | | Cryo-EM structure of SHOC2-PP1c-MRAS holophosphatase complex | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, Leucine-rich repeat protein SHOC-2, ... | | Authors: | Fuller, J.R, Hajian, B, Lemke, C, Kwon, J, Bian, Y, Aguirre, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex.
Nature, 609, 2022
|
|
7UMG
 
 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
1BMA
 
 | | BENZYL METHYL AMINIMIDE INHIBITOR COMPLEXED TO PORCINE PANCREATIC ELASTASE | | Descriptor: | (1R)-1-benzyl-1-methyl-1-(2-{[4-(1-methylethyl)phenyl]amino}-2-oxoethyl)-2-{(2S)-4-methyl-2-[(trifluoroacetyl)amino]pentanoyl}diazanium, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Peisach, E, Casebier, D, Gallion, S.L, Furth, P, Petsko, G.A, Hogan Jr, J.C, Ringe, D. | | Deposit date: | 1995-05-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of a peptidomimetic aminimide inhibitor with elastase.
Science, 269, 1995
|
|
1C6X
 
 | |
1C6Y
 
 | |
7VLI
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2220 | | Descriptor: | 1-[(3~{S},6~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
8JN7
 
 | | Cryo-EM structure of the big tail club shape particle of dengue virus serotype 3 strain CH53489 in complex with human antibody DENV-290 Fab at 37 deg C | | Descriptor: | Human antibody DENV-290 heavy chain, Human antibody DENV-290 light chain, Polyprotein (Fragment) | | Authors: | Fibriansah, G, Ng, T.S, Tan, A.W.K, Shi, J, Lok, S.M. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Ultrapotent human antibodies lock E protein dimers of diverse DENV3 morphological variants
To Be Published
|
|
