5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
3R29
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 | | Descriptor: | Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
3R2A
 
 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 and antagonist rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
1UYT
 
 | | Acetyl-CoA carboxylase carboxyltransferase domain | | Descriptor: | ACETYL-COA CARBOXYLASE | | Authors: | Zhang, H, Tweel, B, Tong, L. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis for the Inhibition of the Carboxyltransferase Domain of Acetyl-Coenzyme-A Carboxylase by Haloxyfop and Diclofop.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
1UYV
 
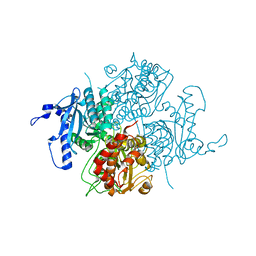 | |
1UYR
 
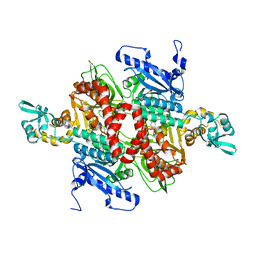 | |
1UYS
 
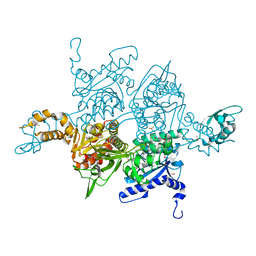 | |
3HI5
 
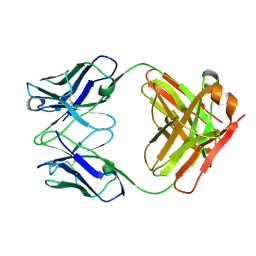 | | Crystal structure of Fab fragment of AL-57 | | Descriptor: | Heavy chain of Fab fragment of AL-57 against alpha L I domain, light chain of Fab fragment of AL-57 against alpha L I domain | | Authors: | Zhang, H, Wang, J. | | Deposit date: | 2009-05-18 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of activation-dependent binding of ligand-mimetic antibody AL-57 to integrin LFA-1.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6CKO
 
 | | Crystal structure of an AF10 fragment | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the DOT1L-AF10 complex reveals mechanistic insights into MLL-AF10-associated leukemogenesis.
Genes Dev., 32, 2018
|
|
3HI6
 
 | |
3OZJ
 
 | | Crystal structure of human retinoic X receptor alpha complexed with bigelovin and coactivator SRC-1 | | Descriptor: | (3aR,4S,4aR,7aR,8R,9aS)-4a,8-dimethyl-3-methylidene-2,5-dioxo-2,3,3a,4,4a,5,7a,8,9,9a-decahydroazuleno[6,5-b]furan-4-yl acetate, Retinoic acid receptor RXR-alpha, SRC-1, ... | | Authors: | Zhang, H, Li, L, Chen, L, Hu, L, Shen, X. | | Deposit date: | 2010-09-25 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure basis of bigelovin as a selective RXR agonist with a distinct binding mode
J.Mol.Biol., 407, 2011
|
|
4E8B
 
 | |
8YR2
 
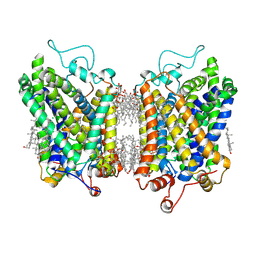 | | Structure of NET-Nisoxetine in outward-open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y92
 
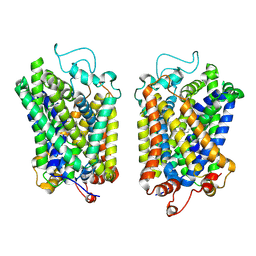 | | structure of NET-Atomoxetine in outward-open state | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y91
 
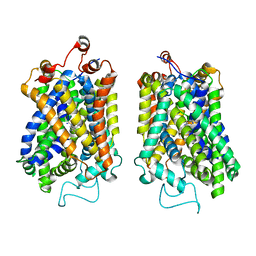 | | Structure of NET-nomifensine in outward-open state | | Descriptor: | (4S)-2-methyl-4-phenyl-3,4-dihydro-1H-isoquinolin-8-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y90
 
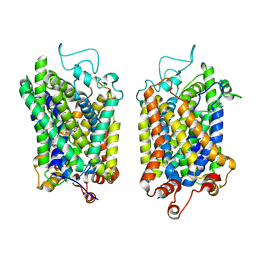 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y93
 
 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y94
 
 | |
8Y8Z
 
 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y95
 
 | | Structure of NET-NE in Occluded state | | Descriptor: | CHLORIDE ION, Noradrenaline, SODIUM ION, ... | | Authors: | Zhang, H, Xu, H.E, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
4KVG
 
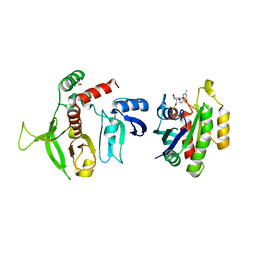 | | Crystal structure of RIAM RA-PH domains in complex with GTP bound Rap1 | | Descriptor: | 1,2-ETHANEDIOL, Amyloid beta A4 precursor protein-binding family B member 1-interacting protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, H, Chang, Y.E, Brennan, M.L, Wu, J. | | Deposit date: | 2013-05-22 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism.
J Mol Cell Biol, 6, 2014
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
4JUR
 
 | |
3RAJ
 
 | |
