3CKD
 
 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3F4A
 
 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3EXM
 
 | | Crystal structure of the phosphatase SC4828 with the non-hydrolyzable nucleotide GPCP | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID GUANOSYL ESTER, ... | | Authors: | Singer, A.U, Xu, X, Zheng, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of a new family of prokaryotic nucleoside diphosphatases.
To be Published
|
|
3EXC
 
 | | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Singer, A.U, Skarina, T, Tan, K, Kagan, O, Onopriyenko, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the RNA'se SSO8090 from Sulfolobus solfataricus
To be Published
|
|
3F4F
 
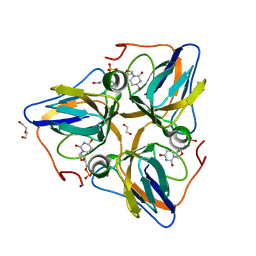 | | Crystal structure of dUT1p, a dUTPase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3HHQ
 
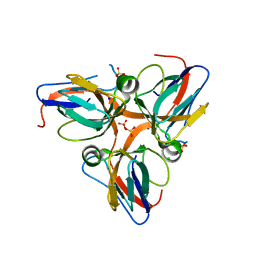 | | Crystal structure of apo dUT1p from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Dong, A, Edwards, A.M, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2009-05-15 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
8T7I
 
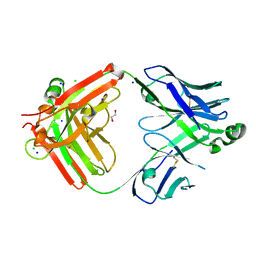 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7G
 
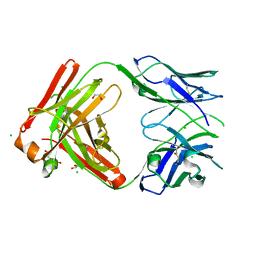 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
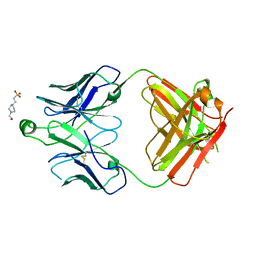 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
3I6Y
 
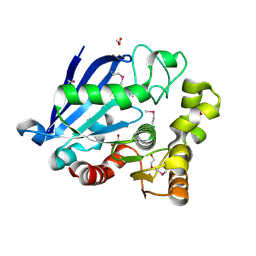 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
8TS5
 
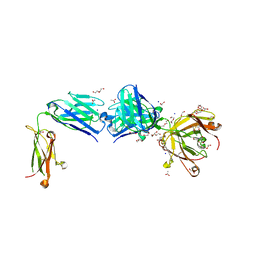 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
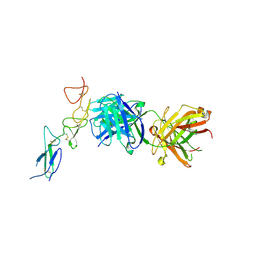 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
3HJZ
 
 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7M3Q
 
 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
4DCA
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound
TO BE PUBLISHED
|
|
7T1L
 
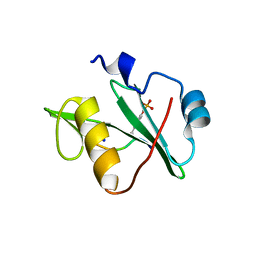 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1K
 
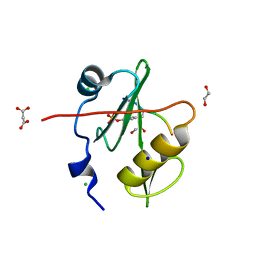 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1U
 
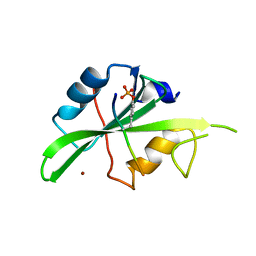 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
1XO5
 
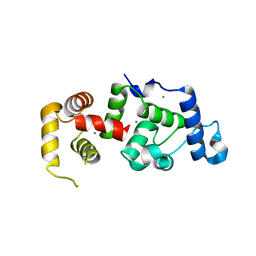 | | Crystal structure of CIB1, an EF-hand, integrin and kinase-binding protein | | Descriptor: | CALCIUM ION, Calcium and integrin-binding protein 1 | | Authors: | Gentry, H.R, Singer, A.U, Betts, L, Yang, C, Ferrara, J.D, Parise, L.V, Sondek, J. | | Deposit date: | 2004-10-05 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Biochemical Characterization of CIB1 Delineates a New Family of EF-hand-containing Proteins
J.Biol.Chem., 280, 2005
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
3P48
 
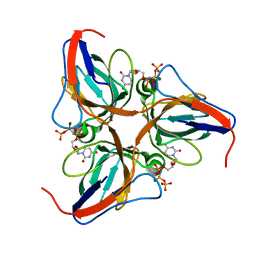 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
2PLE
 
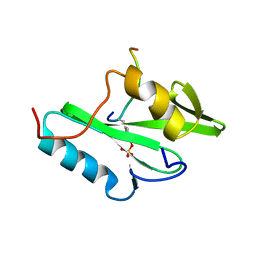 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
2PLD
 
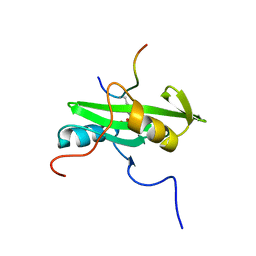 | | NUCLEAR MAGNETIC RESONANCE STRUCTURE OF AN SH2 DOMAIN OF PHOSPHOLIPASE C-GAMMA1 COMPLEXED WITH A HIGH AFFINITY BINDING PEPTIDE | | Descriptor: | PHOSPHOLIPASE C GAMMA-1, C-TERMINAL SH2 DOMAIN, PHOSPHOPEPTIDE FROM PDGF | | Authors: | Pascal, S.M, Singer, A.U, Gish, G, Yamazaki, T, Shoelson, S.E, Pawson, T, Kay, L.E, Forman-Kay, J.D. | | Deposit date: | 1994-08-19 | | Release date: | 1995-01-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of an SH2 domain of phospholipase C-gamma 1 complexed with a high affinity binding peptide.
Cell(Cambridge,Mass.), 77, 1994
|
|
3UZR
 
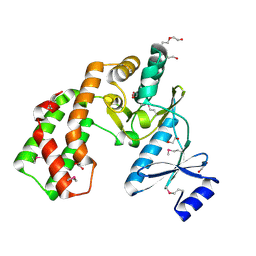 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside phosphotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, apo form
TO BE PUBLISHED
|
|
