7F62
 
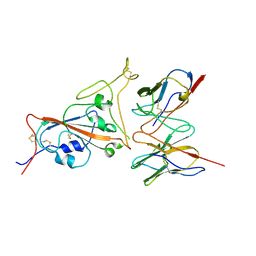 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region) | | Descriptor: | RBD-chAb-25, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
4UHB
 
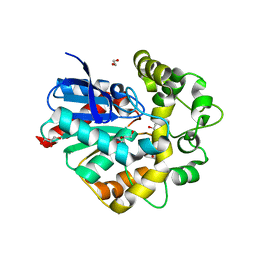 | | Laboratory evolved variant R-C1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,2-ETHANEDIOL, EPOXIDE HYDROLASE, GLYCEROL | | Authors: | Nilsson, M.T.I, Carlsson, A.J, Dobritzsch, D, Widersten, M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
3QT0
 
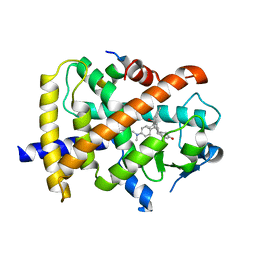 | | Revealing a steroid receptor ligand as a unique PPARgamma agonist | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Nuclear receptor coactivator 1 peptide, Peroxisome proliferator-activated receptor gamma | | Authors: | Rong, H. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Cell Res., 22, 2012
|
|
2POI
 
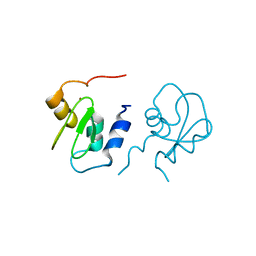 | | Crystal structure of XIAP BIR1 domain (I222 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Lin, S. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCT
 
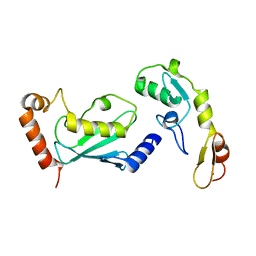 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6C7H
 
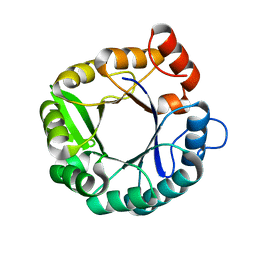 | |
6C7M
 
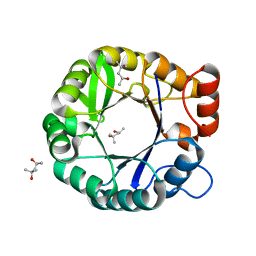 | |
6C7V
 
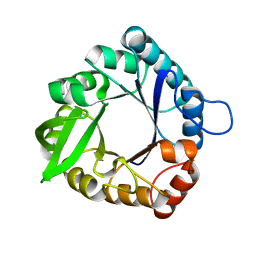 | |
6C8B
 
 | |
6C7T
 
 | |
6CAI
 
 | |
6CT3
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 27 round 7-2 | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, GLYCEROL, Kemp eliminase, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6DKV
 
 | |
6DC1
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 25 round 7 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-nitro-2-oxidanyl-benzenecarbonitrile, ... | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
6DNJ
 
 | | Directed evolutionary changes in Kemp Eliminase KE07 - Crystal 28 round 5 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-nitro-1,2-benzoxazole, Kemp eliminase KE07 | | Authors: | Jackson, C.J, Hong, N.-S, Carr, P.D. | | Deposit date: | 2018-06-06 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The evolution of multiple active site configurations in a designed enzyme.
Nat Commun, 9, 2018
|
|
4Y9S
 
 | |
6BM9
 
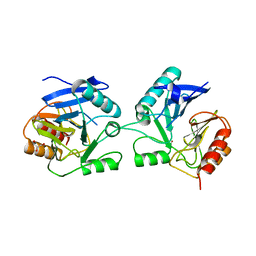 | |
4Q1U
 
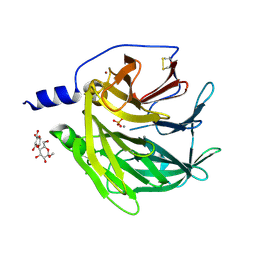 | | Serum paraoxonase-1 by directed evolution with the K192Q mutation | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Catalytic stimulation by restrained active-site floppiness-the case of high density lipoprotein-bound serum paraoxonase-1.
J.Mol.Biol., 427, 2015
|
|
6PYP
 
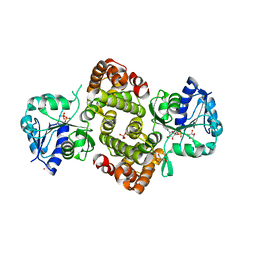 | | Binary Complex of Human Glycerol 3-Phosphate Dehydrogenase, R269A mutant | | Descriptor: | 2,2-bis(hydroxymethyl)propane-1,3-diol, Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic, ... | | Authors: | Gulick, A.M. | | Deposit date: | 2019-07-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modeling the Role of a Flexible Loop and Active Site Side Chains in Hydride Transfer Catalyzed by Glycerol-3-phosphate Dehydrogenase.
Acs Catalysis, 10, 2020
|
|
7S4F
 
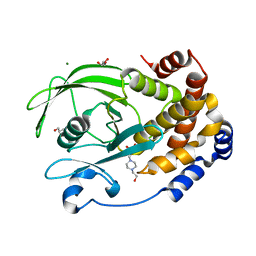 | | Protein Tyrosine Phosphatase 1B - F182Q mutant bound with Hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brandao, T.A.S, Hengge, A.C, Johnson, S.J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
430D
 
 | | STRUCTURE OF SARCIN/RICIN LOOP FROM RAT 28S RRNA | | Descriptor: | MAGNESIUM ION, SARCIN/RICIN LOOP FROM RAT 28S R-RNA | | Authors: | Correll, C.C, Munishkin, A, Chan, Y.L, Ren, Z, Wool, I.G, Steitz, T.A. | | Deposit date: | 1998-10-04 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ribosomal RNA domain essential for binding elongation factors.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4UHU
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase class A | | Descriptor: | ACETATE ION, FORMIC ACID, GNCA LACTAMASE W229D | | Authors: | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5AJ9
 
 | | G7 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ARYLSULFATASE, CALCIUM ION, ... | | Authors: | Miton, C.M, Fischer, G, Jonas, S, Mohammed, M.F, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
